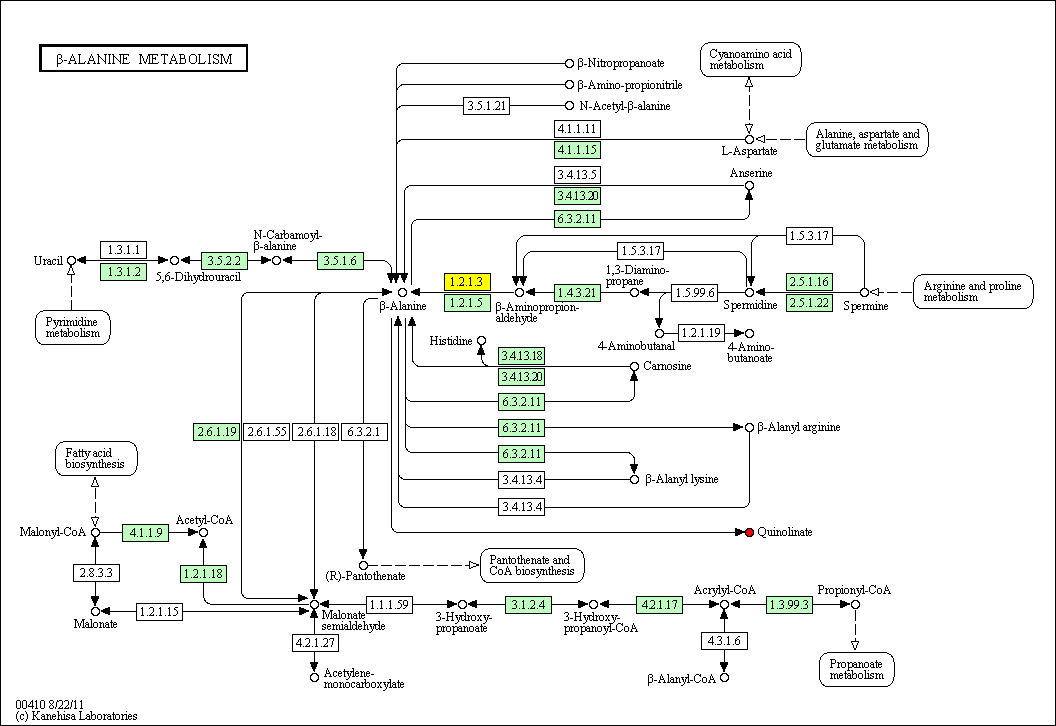
cpd:C00018 Pyridoxal phosphate
cpd:C00019 S-Adenosyl-L-methionine
cpd:C00025 L-Glutamate
cpd:C00031 D-Glucose
cpd:C00051 Glutathione
cpd:C00072 Ascorbate
cpd:C00078 L-Tryptophan
cpd:C00155 L-Homocysteine
cpd:C00187 Cholesterol
cpd:C00250 Pyridoxal
cpd:C00253 Nicotinate
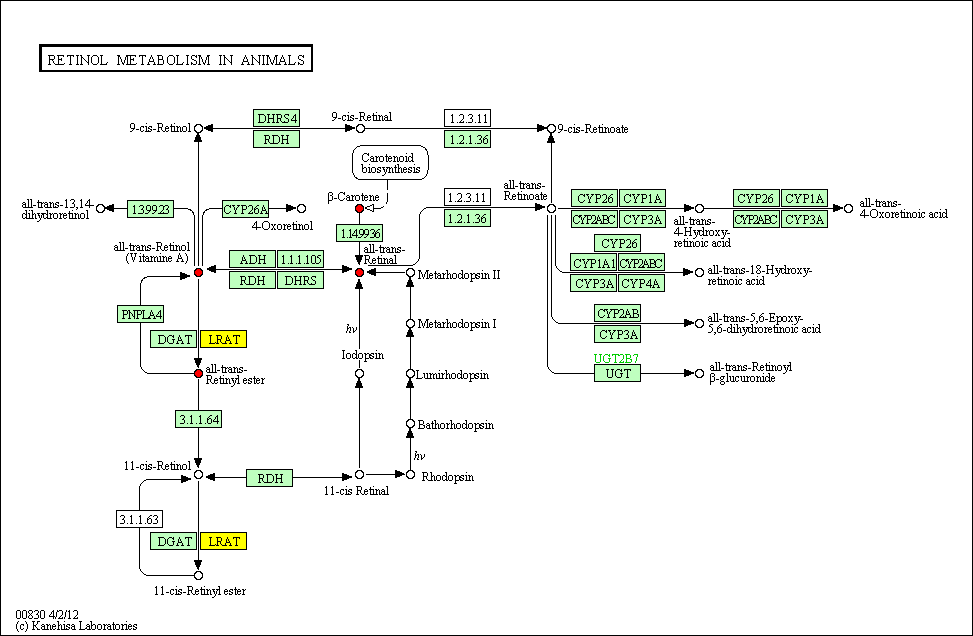
cpd:C00376 Retinal
cpd:C00378 Thiamine
cpd:C00422 Triacylglycerol
cpd:C00473 Retinol
cpd:C00504 Folate
cpd:C00534 Pyridoxamine
cpd:C00535 Testosterone
cpd:C00951 Estradiol-17beta
cpd:C01717 4-Hydroxy-2-quinolinecarboxylic acid
cpd:C02094 beta-Carotene
cpd:C02477 alpha-Tocopherol
cpd:C03722 Quinolinate
cpd:C05441 Vitamin D2
cpd:C05443 Vitamin D3
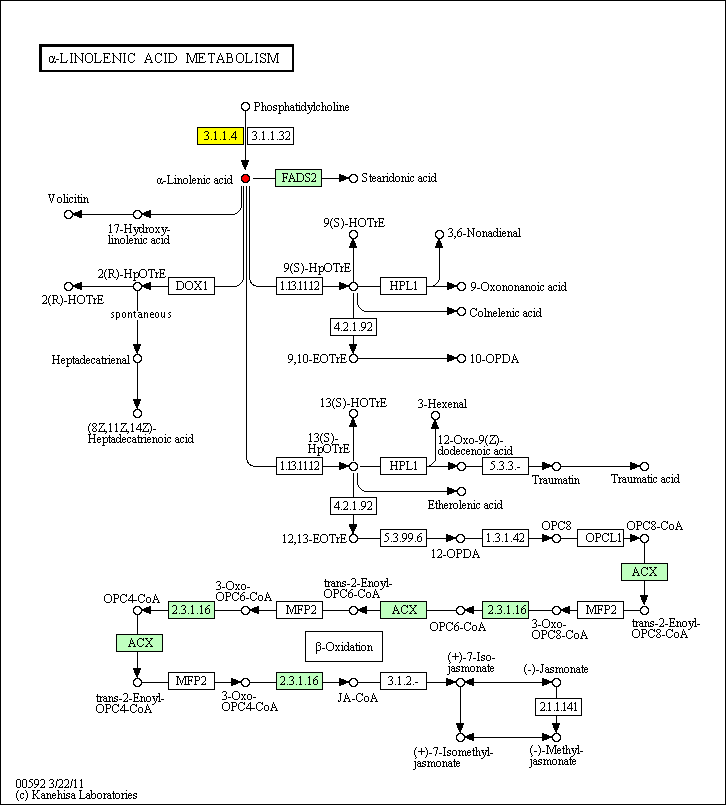
cpd:C06427 (9Z,12Z,15Z)-Octadecatrienoic acid
hsa:27034 ACAD8; acyl-CoA dehydrogenase family, member 8
hsa:10554 AGPAT1; 1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (EC:2.3.1.51)
hsa:5832 ALDH18A1; aldehyde dehydrogenase 18 family, member A1 (EC:1.2.1.41 2.7.2.11)
hsa:217 ALDH2; aldehyde dehydrogenase 2 family (mitochondrial) (EC:1.2.1.3)
hsa:411 ARSB; arylsulfatase B (EC:3.1.6.12)
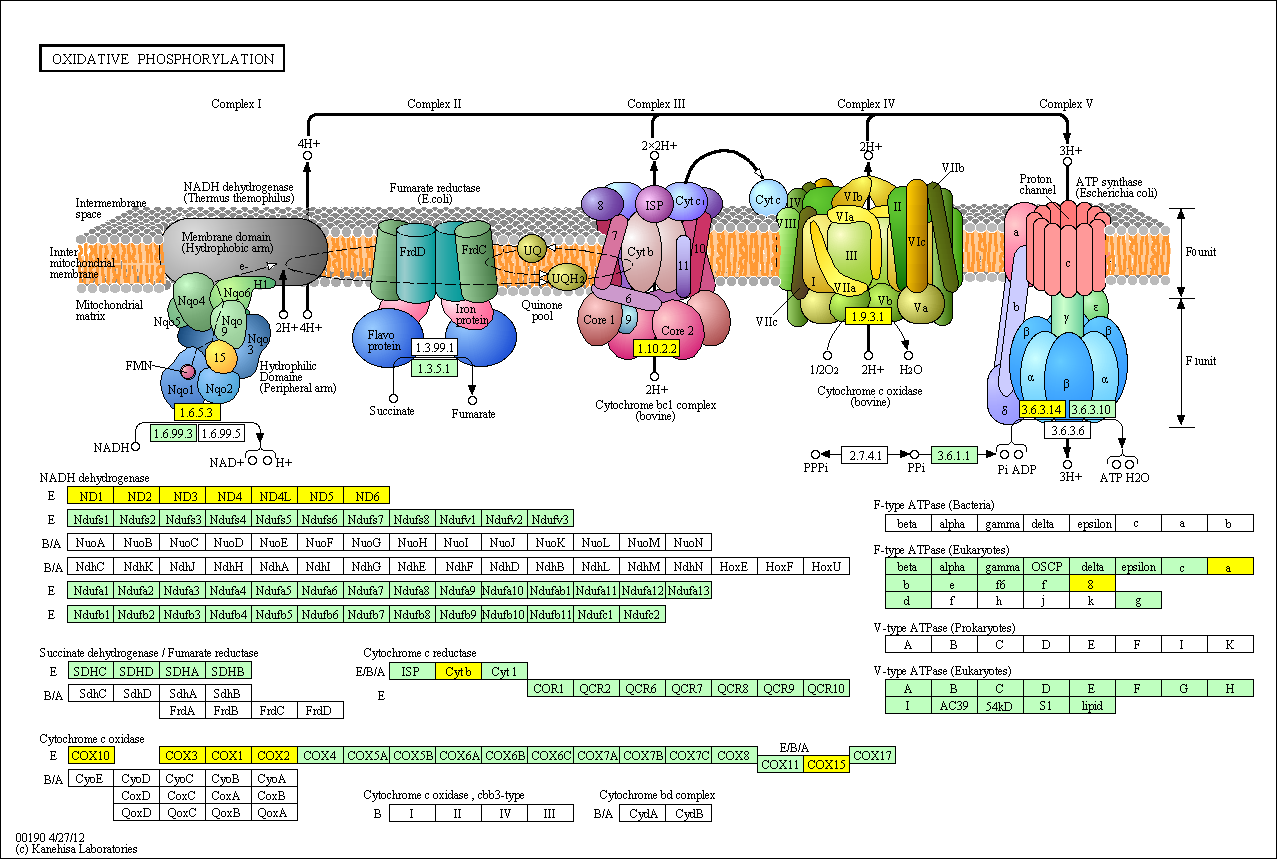
hsa:4508 ATP6; ATP synthase F0 subunit 6
hsa:4509 ATP8; ATP synthase F0 subunit 8
hsa:875 CBS; cystathionine-beta-synthase (EC:4.2.1.22)
hsa:1312 COMT; catechol-O-methyltransferase (EC:2.1.1.6)
hsa:4512 COX1; cytochrome c oxidase subunit I
hsa:1352 COX10; COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast) (EC:2.5.1.-)
hsa:1355 COX15; COX15 homolog, cytochrome c oxidase assembly protein (yeast)
hsa:4513 COX2; cytochrome c oxidase subunit II
hsa:4514 COX3; cytochrome c oxidase III
hsa:1588 CYP19A1; cytochrome P450, family 19, subfamily A, polypeptide 1 (EC:1.14.14.1)
hsa:4519 CYTB; cytochrome b
hsa:1621 DBH; dopamine beta-hydroxylase (dopamine beta-monooxygenase) (EC:1.14.17.1)
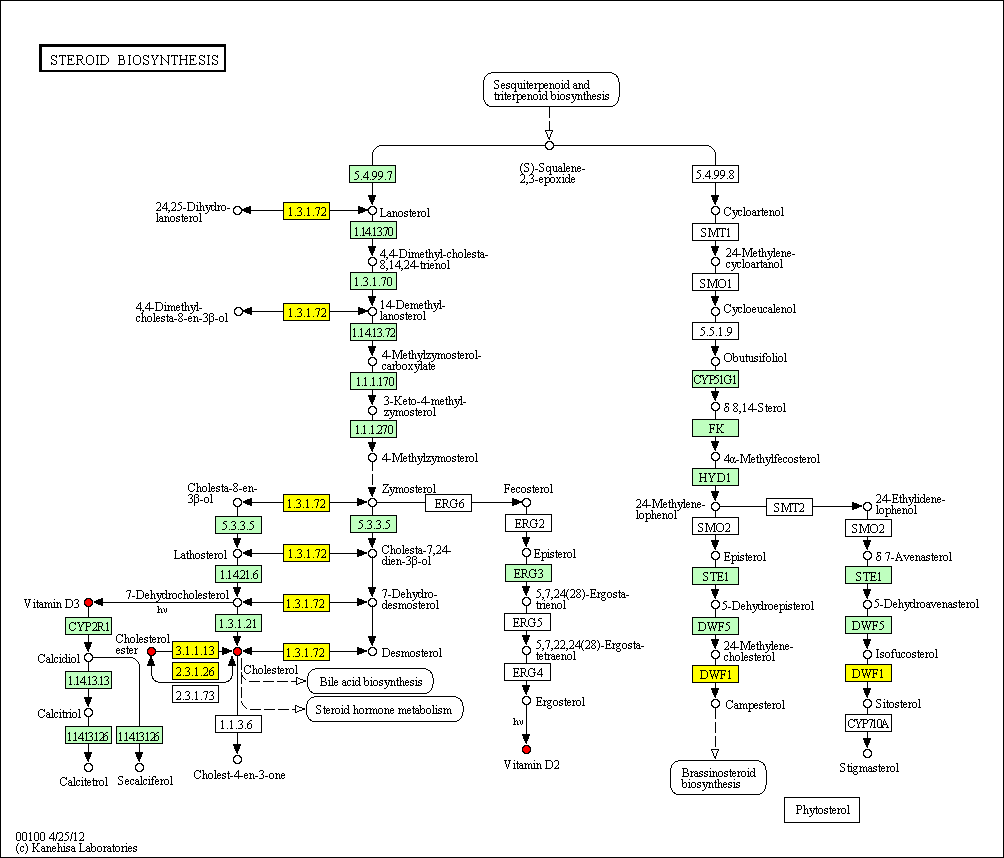
hsa:1718 DHCR24; 24-dehydrocholesterol reductase (EC:1.3.1.72)
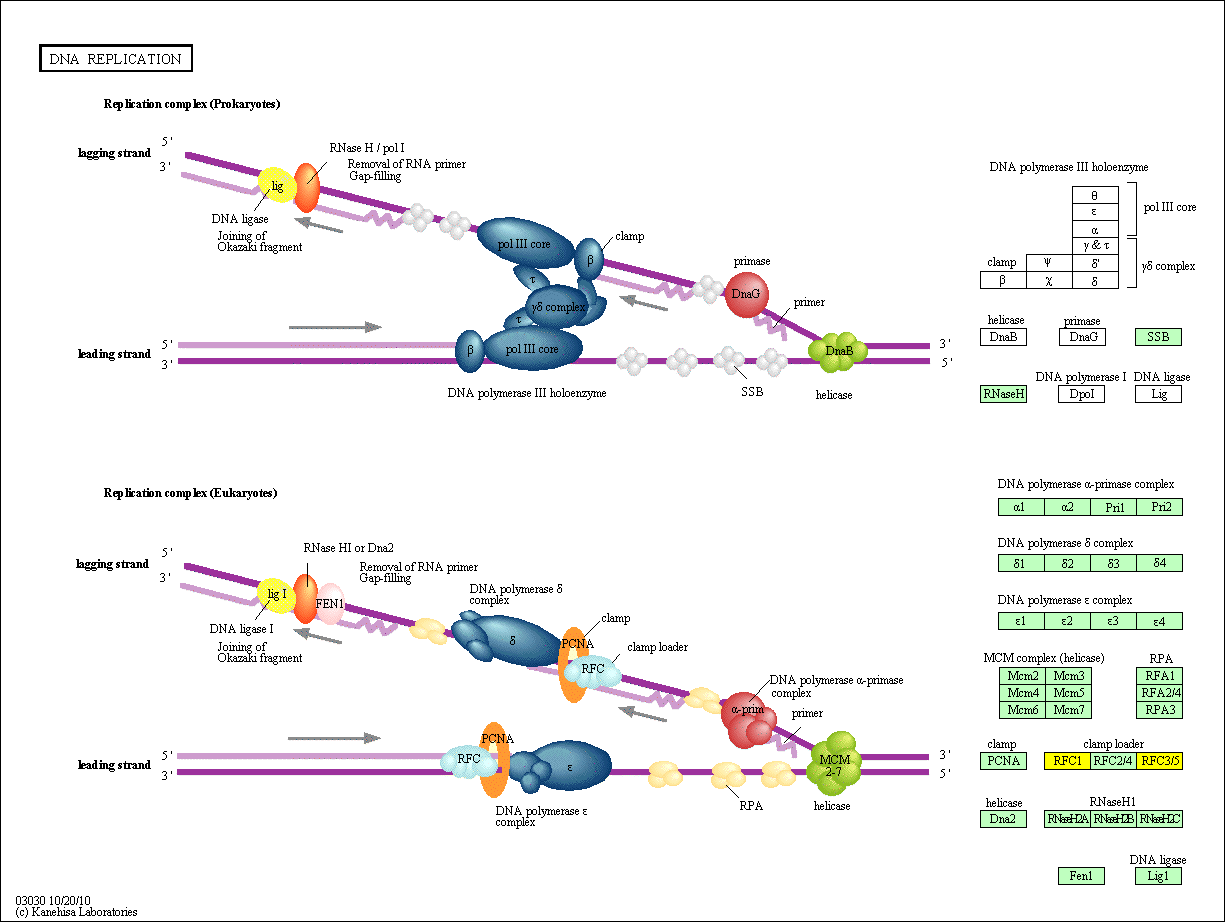
hsa:1738 DLD; dihydrolipoamide dehydrogenase (EC:1.8.1.4)
hsa:1743 DLST; dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) (EC:2.3.1.61)
hsa:2224 FDPS; farnesyl diphosphate synthase (EC:2.5.1.1 2.5.1.10)
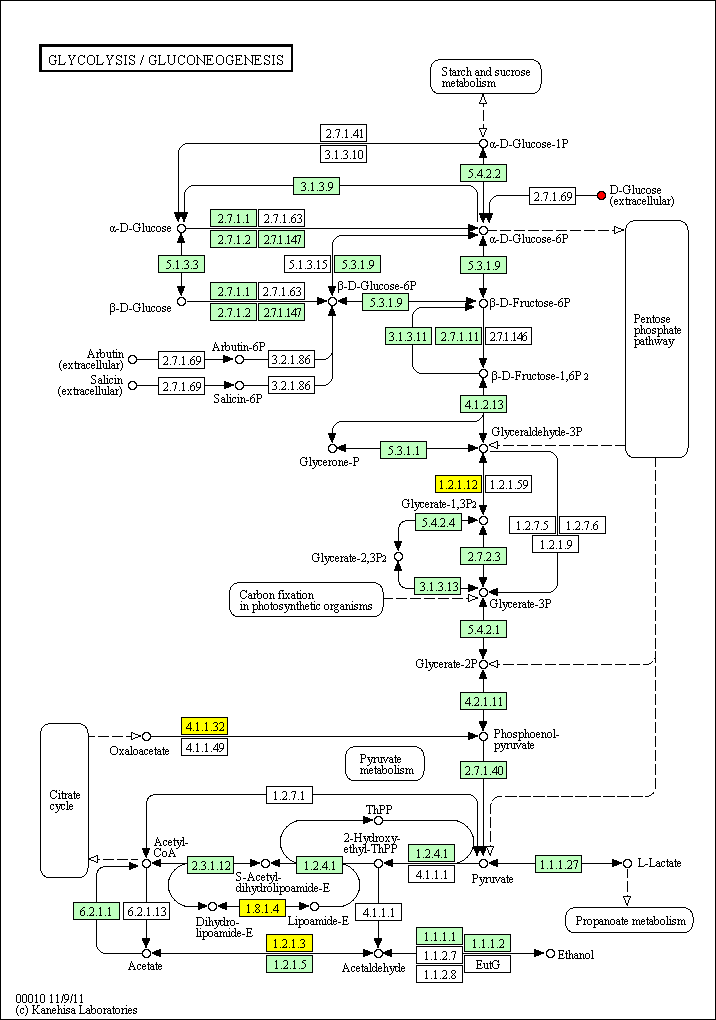
hsa:2597 GAPDH; glyceraldehyde-3-phosphate dehydrogenase (EC:1.2.1.12)
hsa:26330 GAPDHS; glyceraldehyde-3-phosphate dehydrogenase, spermatogenic (EC:1.2.1.12)
hsa:3156 HMGCR; 3-hydroxy-3-methylglutaryl-CoA reductase (EC:1.1.1.34)
hsa:3158 HMGCS2; 3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) (EC:2.3.3.10)
hsa:3290 HSD11B1; hydroxysteroid (11-beta) dehydrogenase 1 (EC:1.1.1.146)
hsa:3990 LIPC; lipase, hepatic (EC:3.1.1.3)
hsa:4128 MAOA; monoamine oxidase A (EC:1.4.3.4)
hsa:25902 MTHFD1L; methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like (EC:6.3.4.3)
hsa:4524 MTHFR; methylenetetrahydrofolate reductase (NAD(P)H) (EC:1.5.1.20)
hsa:4548 MTR; 5-methyltetrahydrofolate-homocysteine methyltransferase (EC:2.1.1.13)
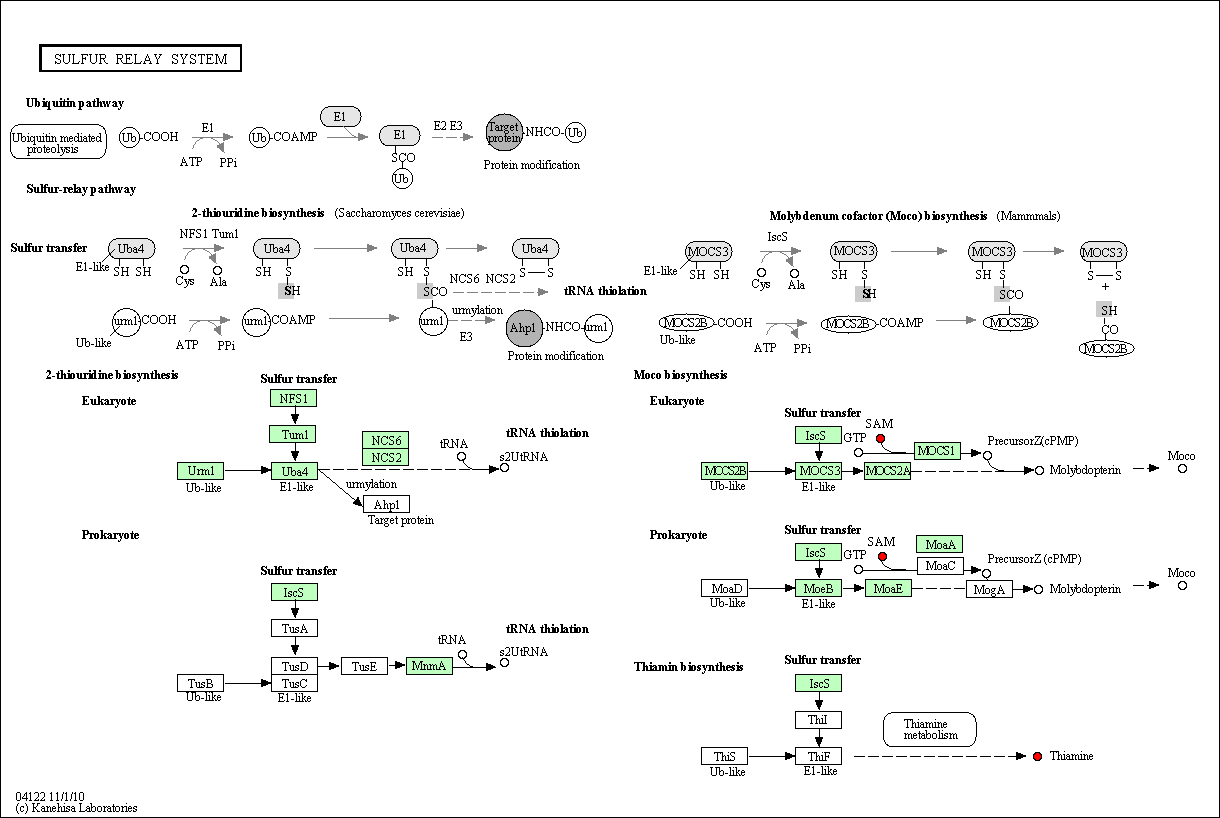
MTRR 5-methyltetrahydrofolate-homocysteine methyltransferase reductase belongs here
hsa:10 NAT2; N-acetyltransferase 2 (arylamine N-acetyltransferase) (EC:2.3.1.5)
hsa:4535 ND1; NADH dehydrogenase, subunit 1 (complex I)
hsa:4536 ND2; MTND2
hsa:4537 ND3; NADH dehydrogenase, subunit 3 (complex I)
hsa:4538 ND4; NADH dehydrogenase, subunit 4 (complex I)
hsa:4539 ND4L; NADH dehydrogenase, subunit 4L (complex I)
hsa:4540 ND5; NADH dehydrogenase, subunit 5 (complex I)
hsa:4541 ND6; NADH dehydrogenase, subunit 6 (complex I)
hsa:349565 NMNAT3; nicotinamide nucleotide adenylyltransferase 3 (EC:2.7.7.1 2.7.7.18)
hsa:4842 NOS1; nitric oxide synthase 1 (neuronal) (EC:1.14.13.39)
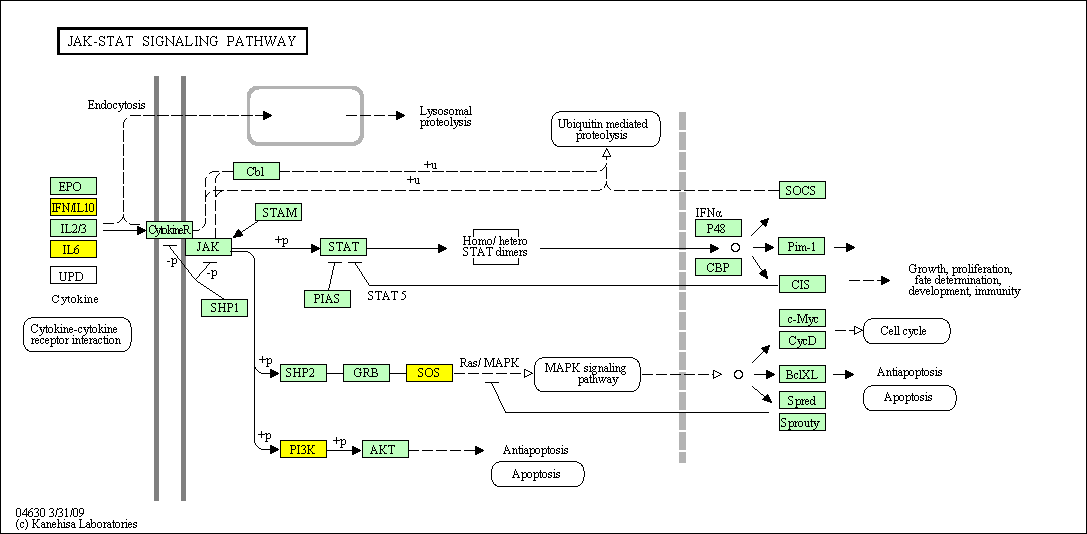
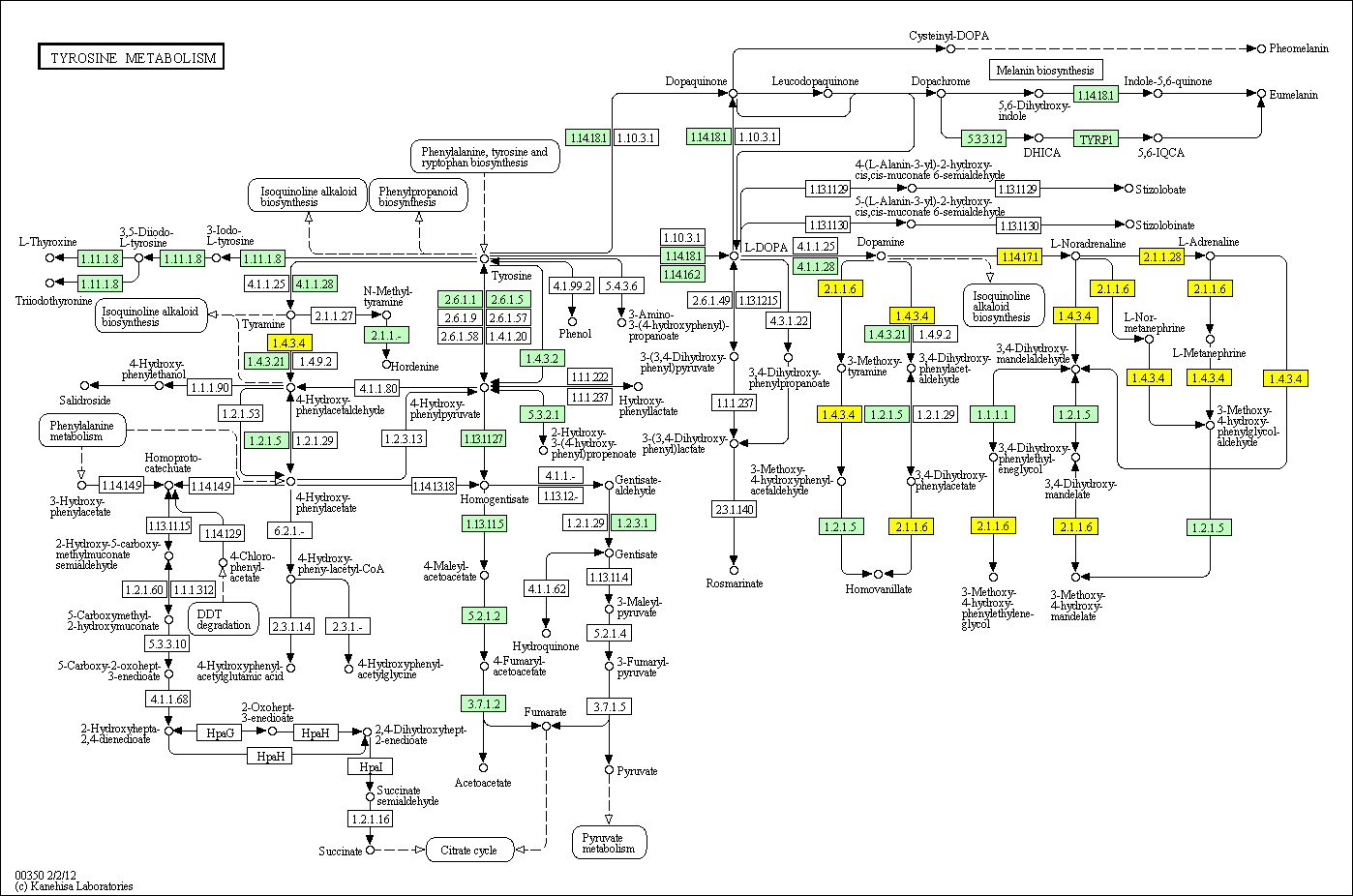
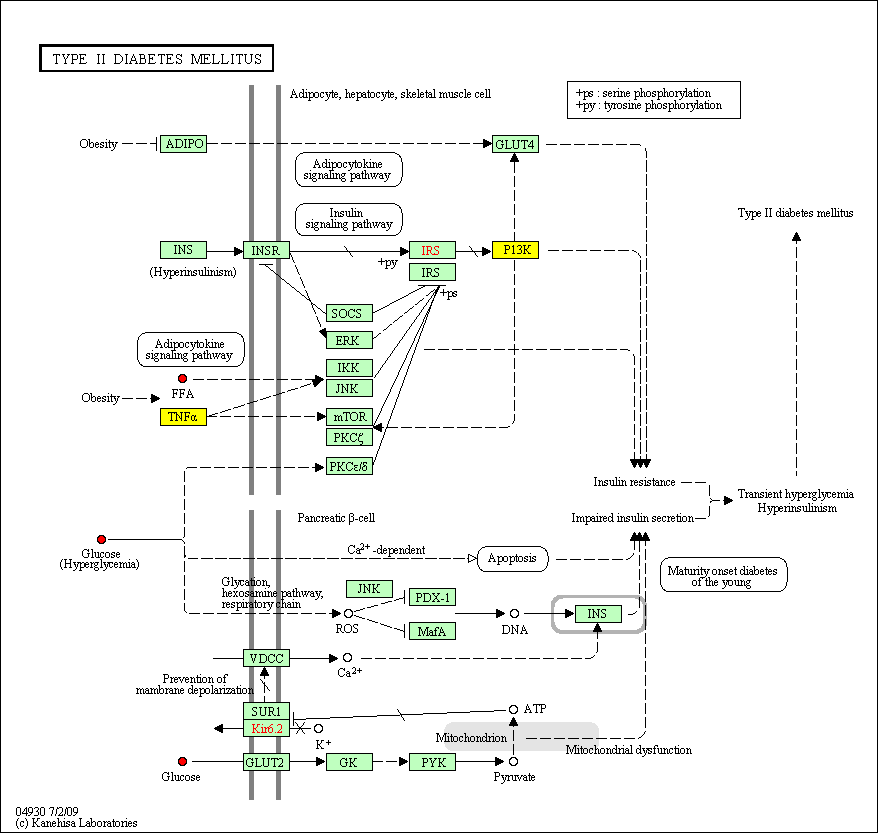
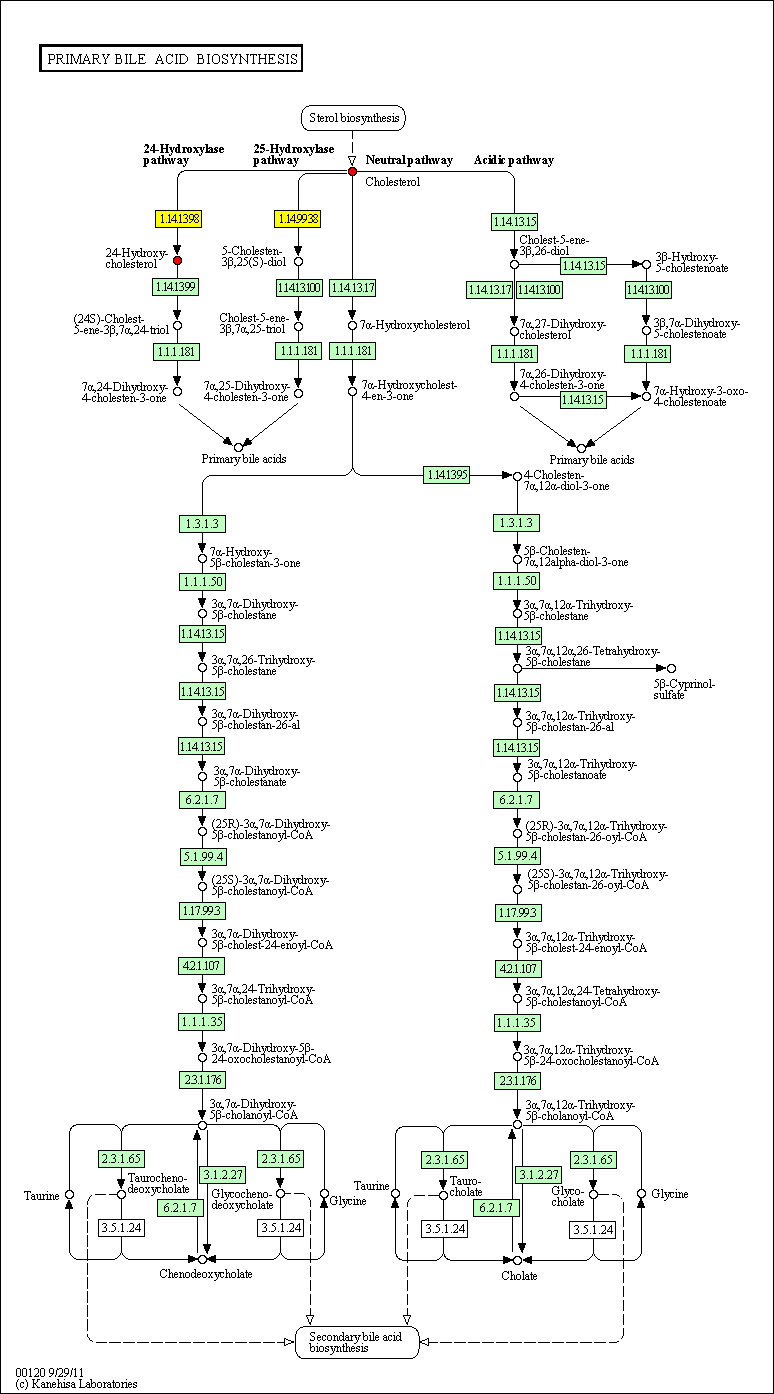
hsa:4843 NOS2; nitric oxide synthase 2, inducible (EC:1.14.13.39)
hsa:4846 NOS3; nitric oxide synthase 3 (endothelial cell) (EC:1.14.13.39)
hsa:5009 OTC; ornithine carbamoyltransferase (EC:2.1.3.3)
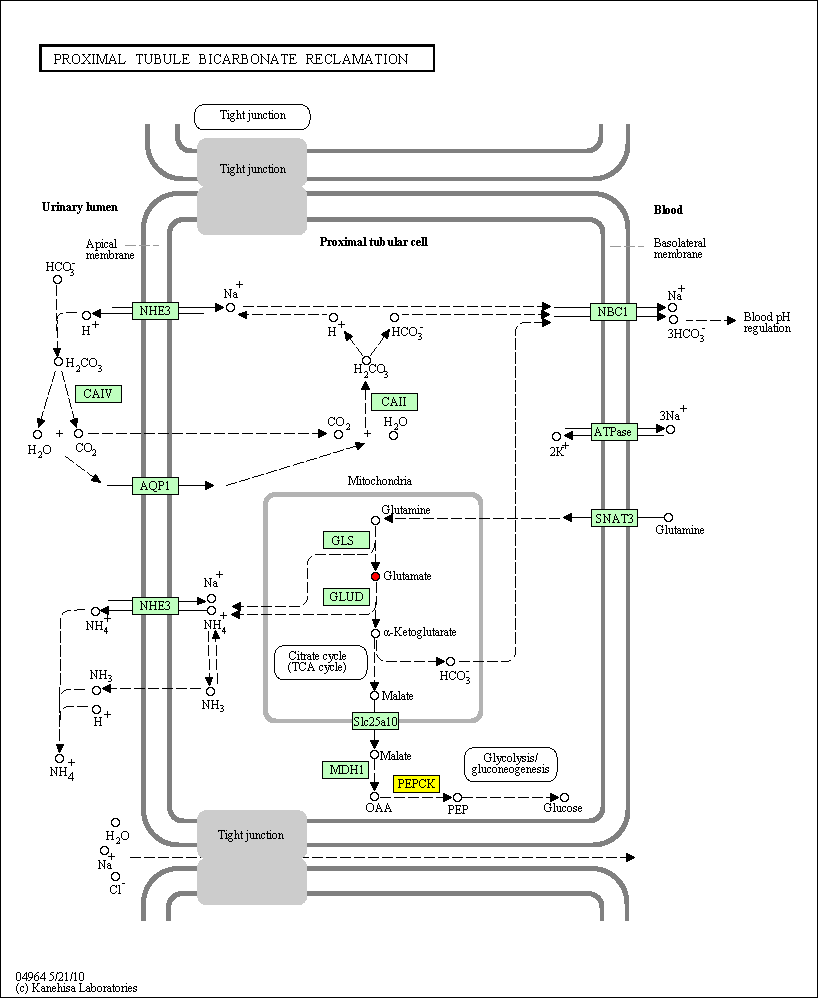
hsa:5105 PCK1; phosphoenolpyruvate carboxykinase 1 (soluble) (EC:4.1.1.32)
hsa:50487 PLA2G3; phospholipase A2, group III (EC:3.1.1.4)
hsa:5409 PNMT; phenylethanolamine N-methyltransferase (EC:2.1.1.28)
hsa:5444 PON1; paraoxonase 1 (EC:3.1.1.2 3.1.8.1 3.1.1.81)
hsa:5445 PON2; paraoxonase 2 (EC:3.1.1.2 3.1.1.81)
hsa:5446 PON3; paraoxonase 3 (EC:3.1.1.2 3.1.8.1 3.1.1.81)
hsa:5743 PTGS2; prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1)
hsa:8879 SGPL1; sphingosine-1-phosphate lyase 1 (EC:4.1.2.27)
hsa:66008 TRAK2; trafficking protein, kinesin binding 2
hsa:64409 WBSCR17; Williams-Beuren syndrome chromosome region 17 (EC:2.4.1.41)
 Subscribe
to RSS feed
Subscribe
to RSS feed


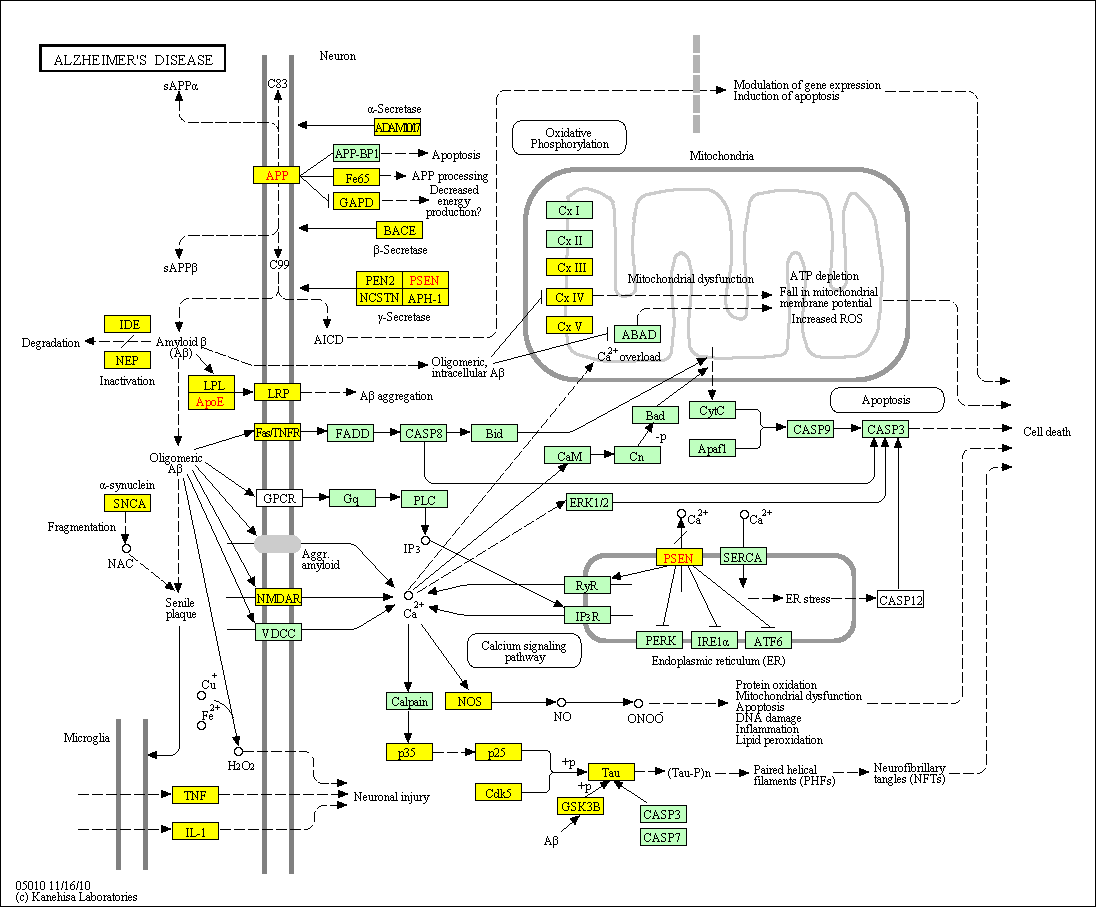
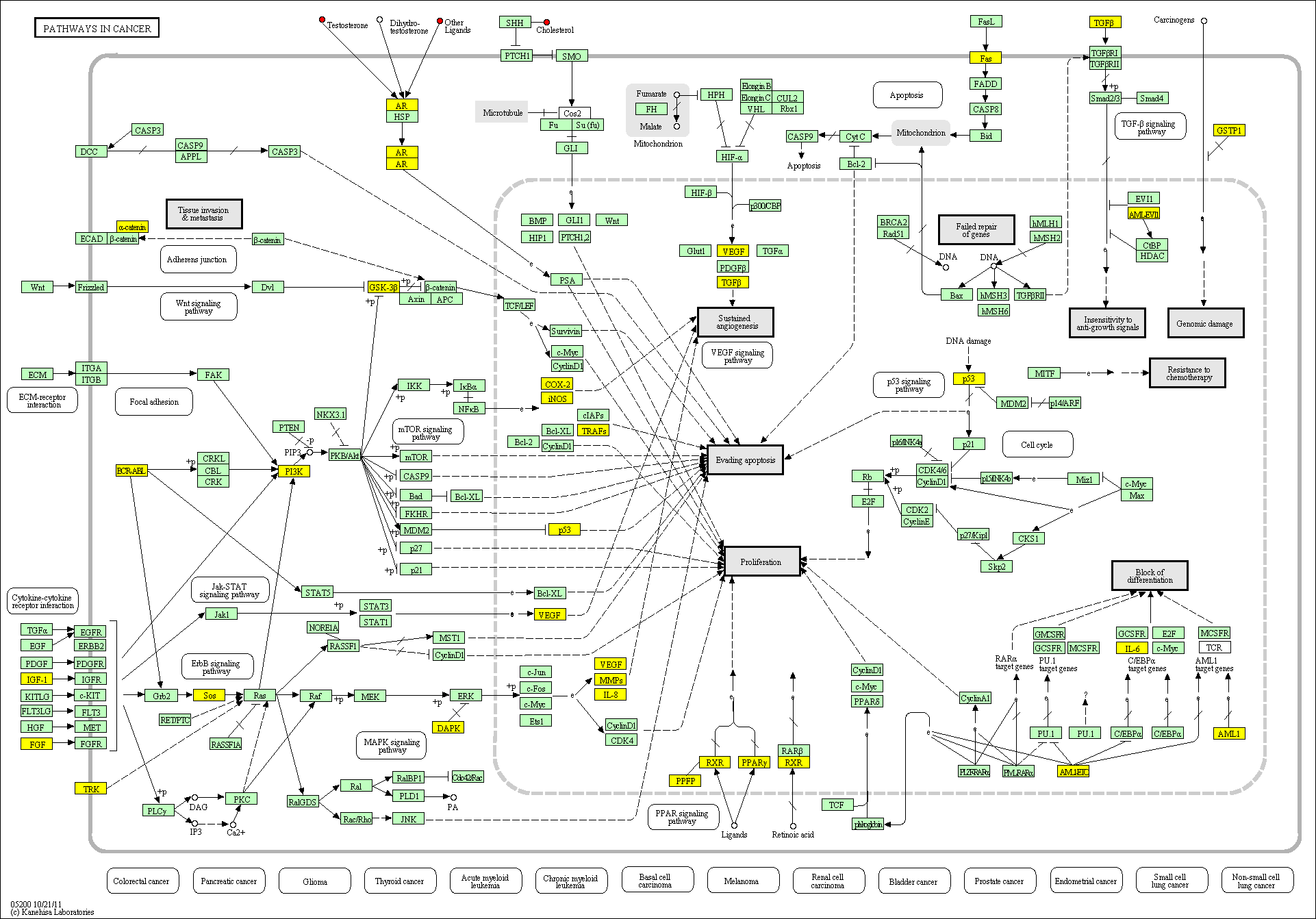
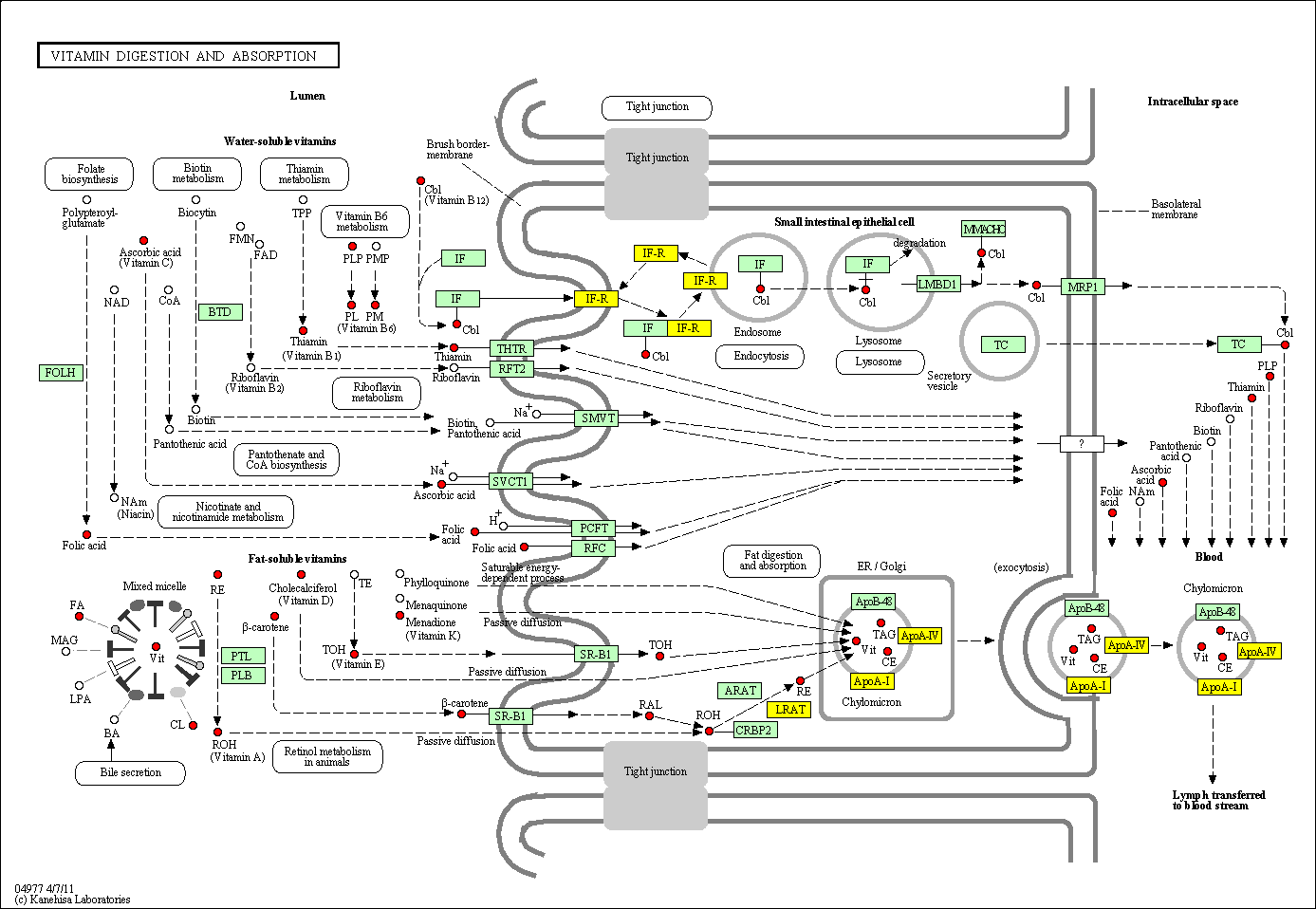
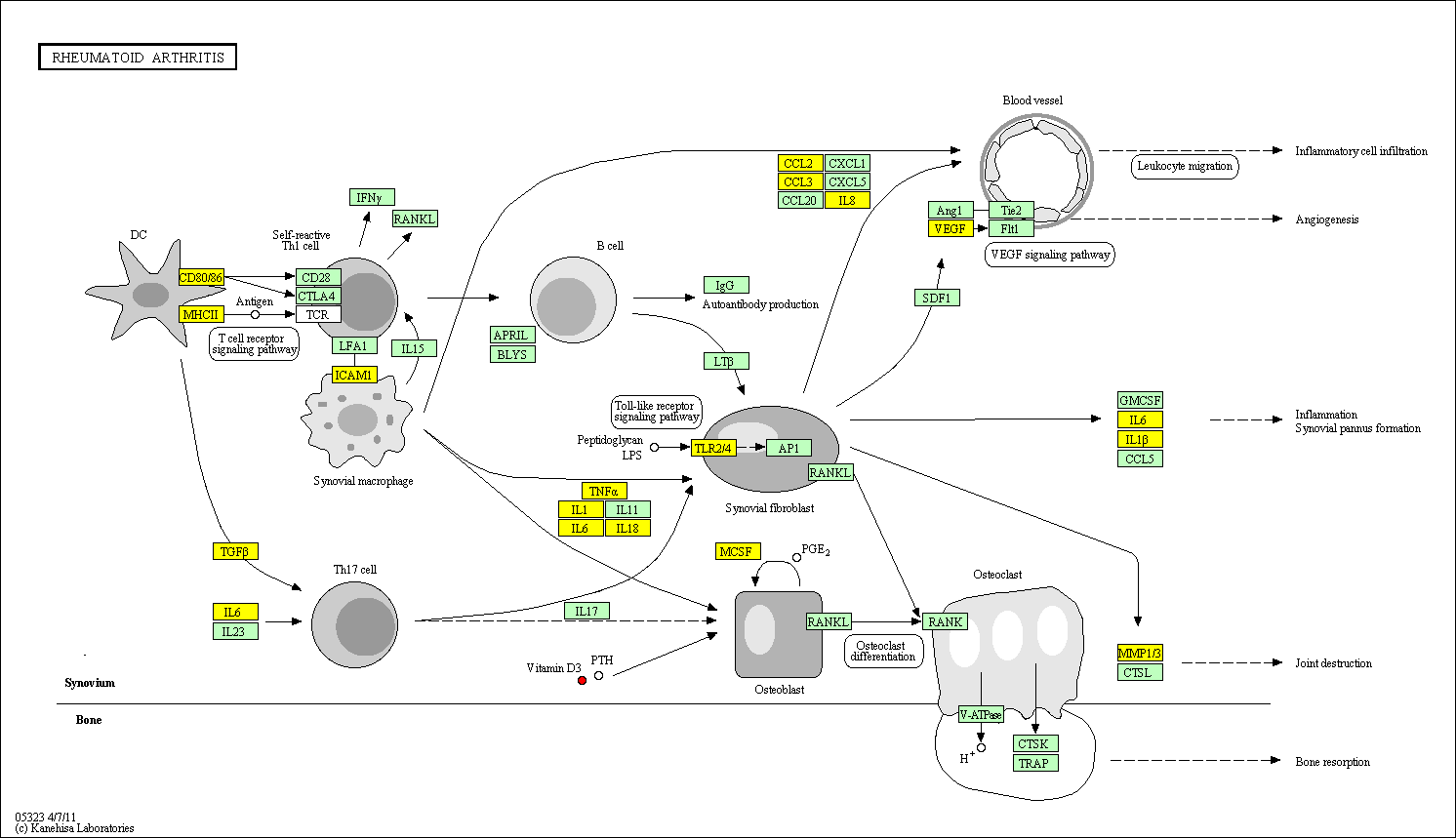
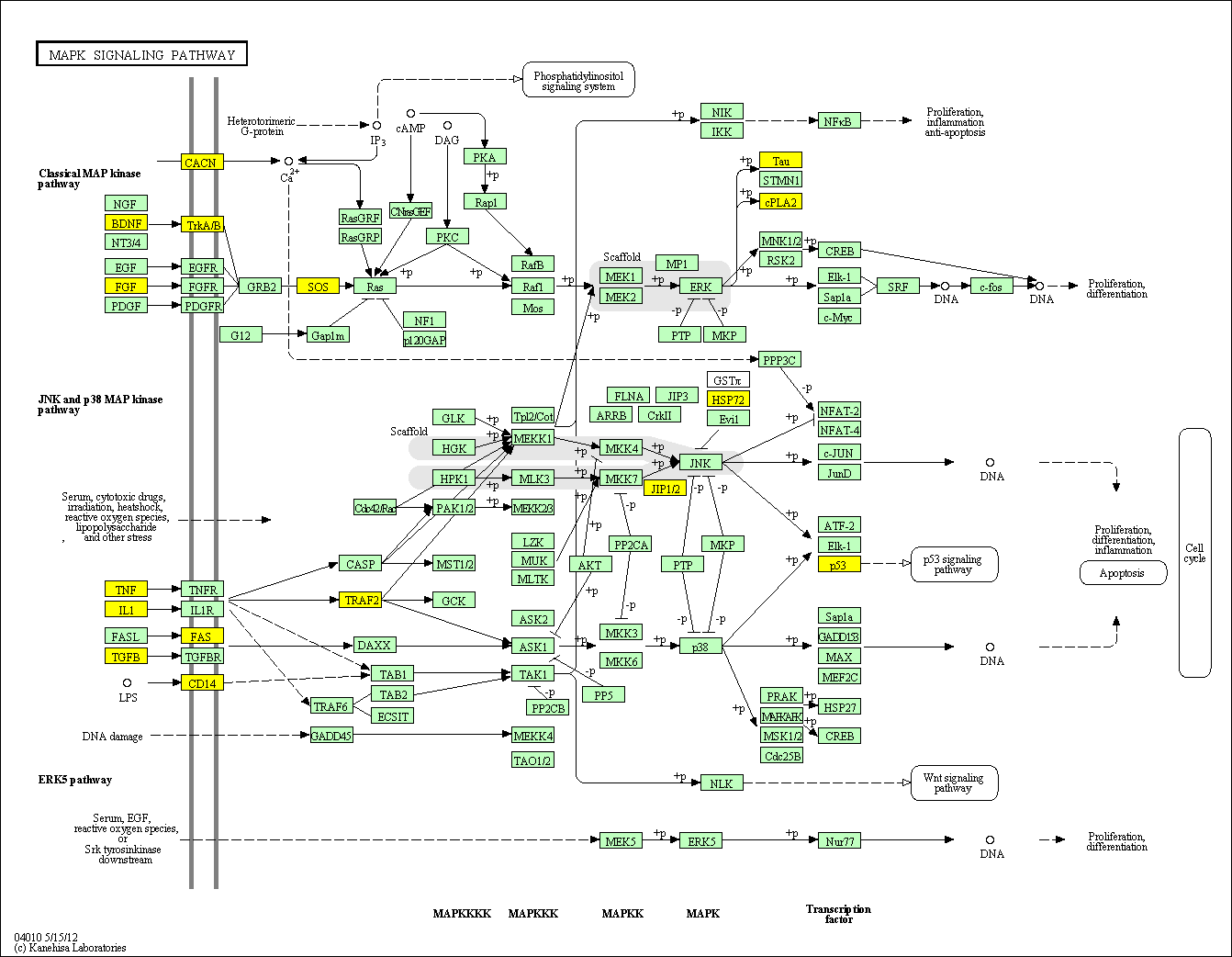
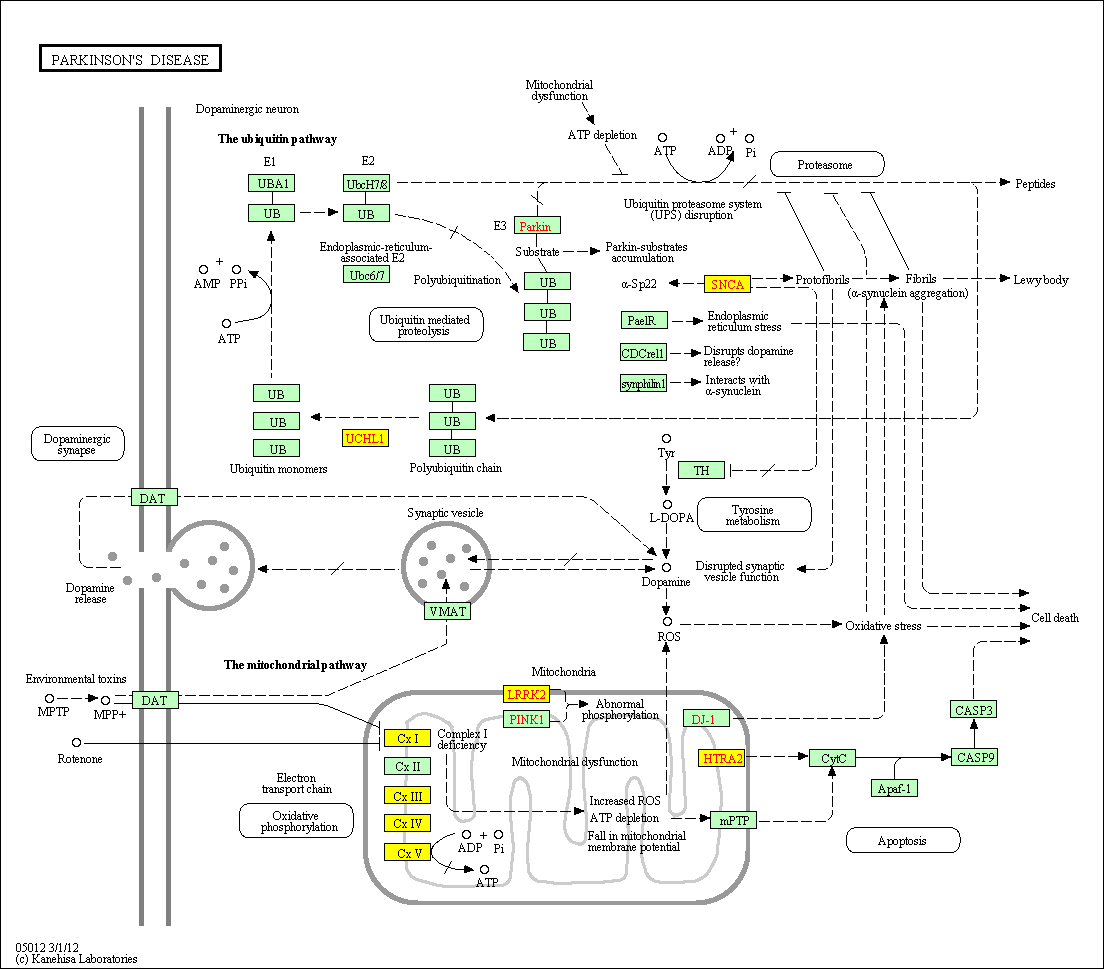
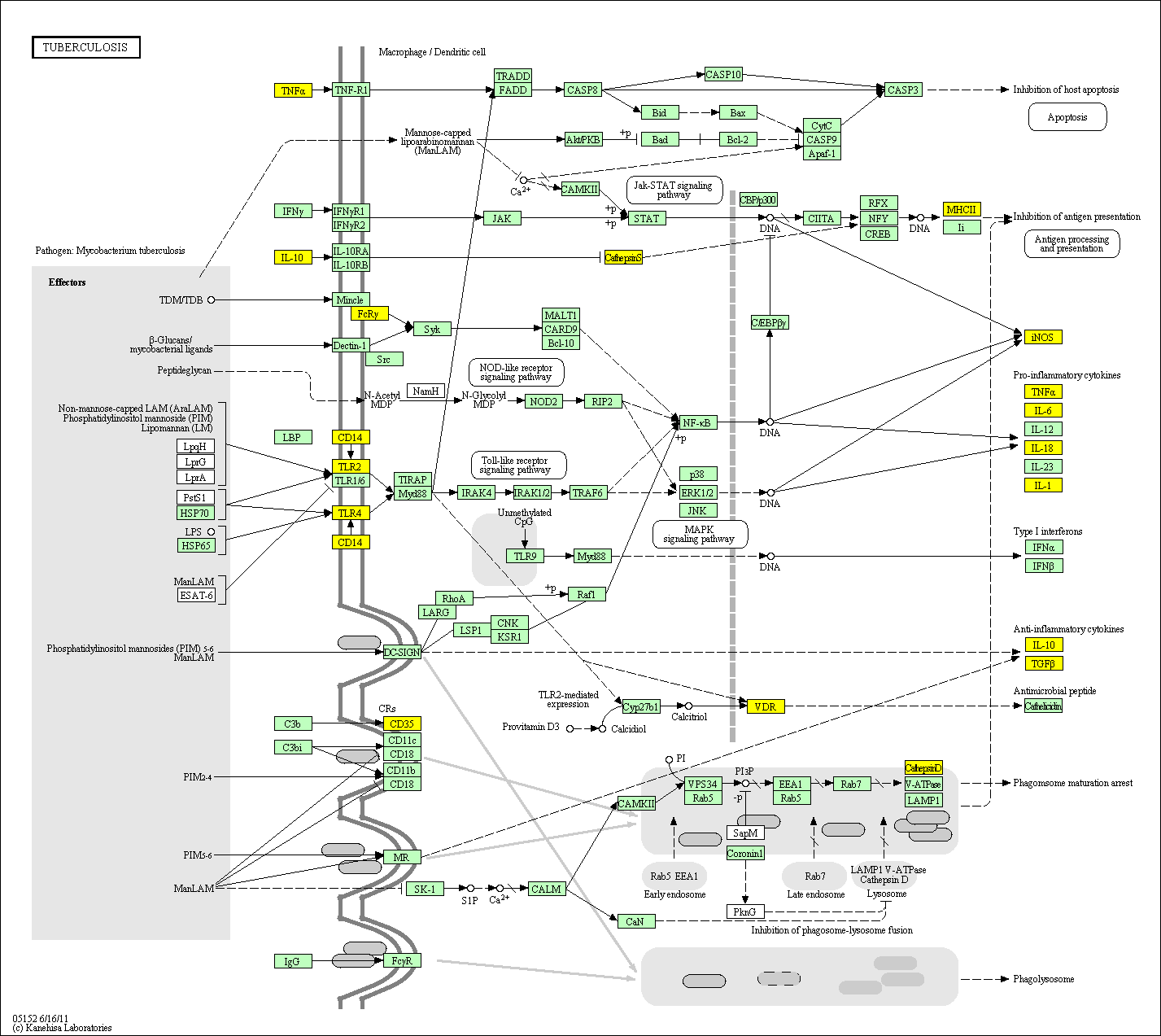
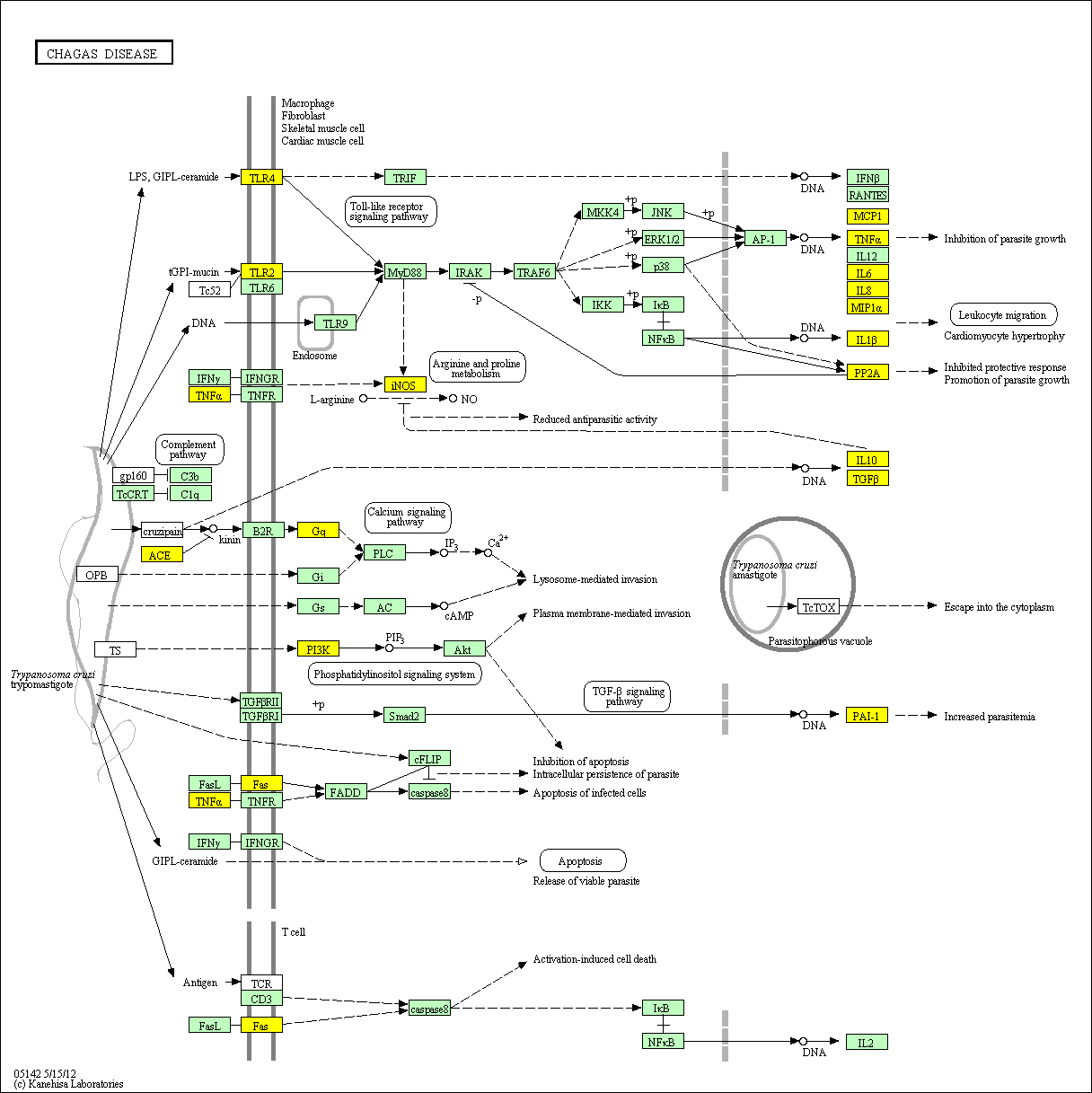
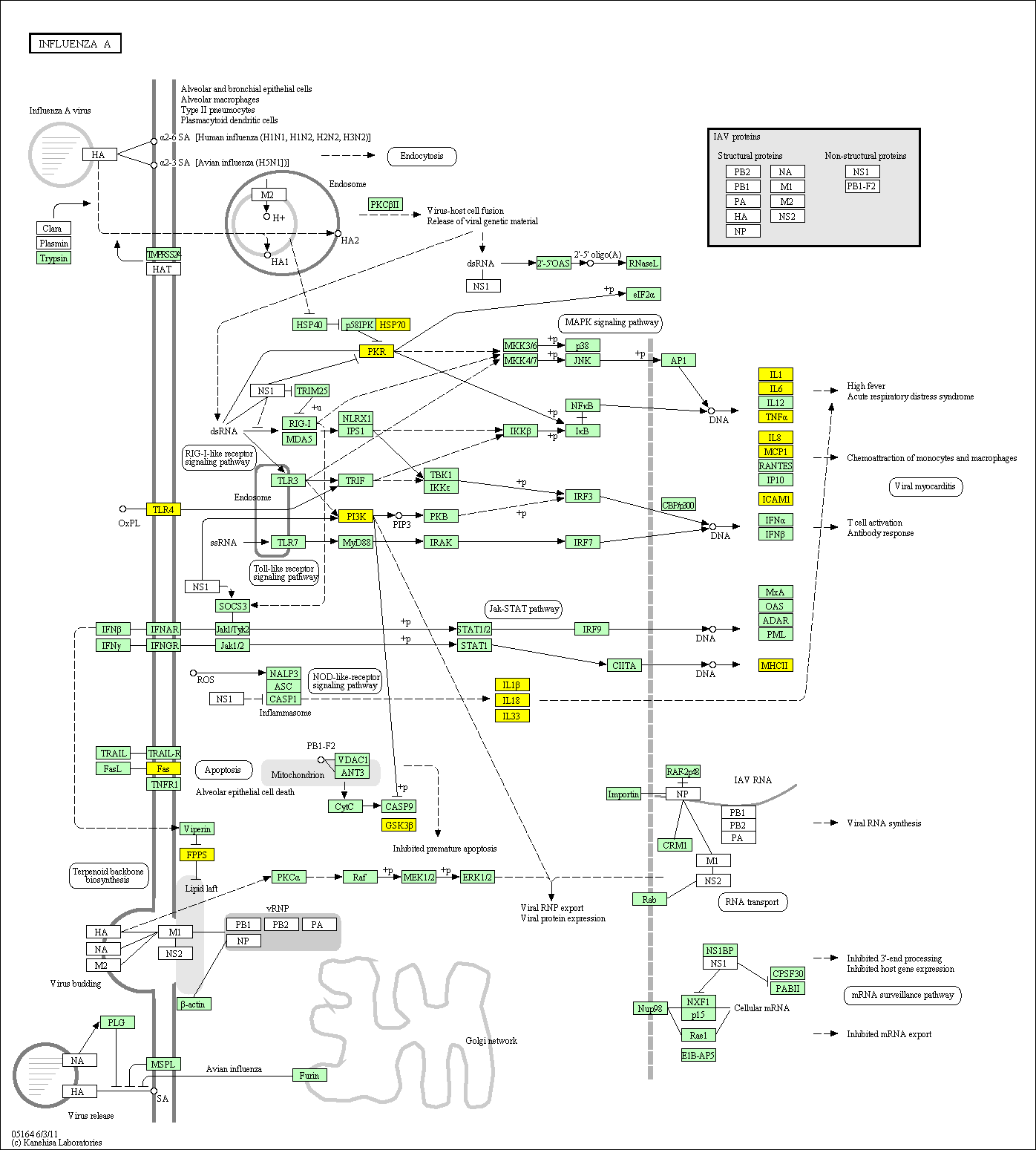
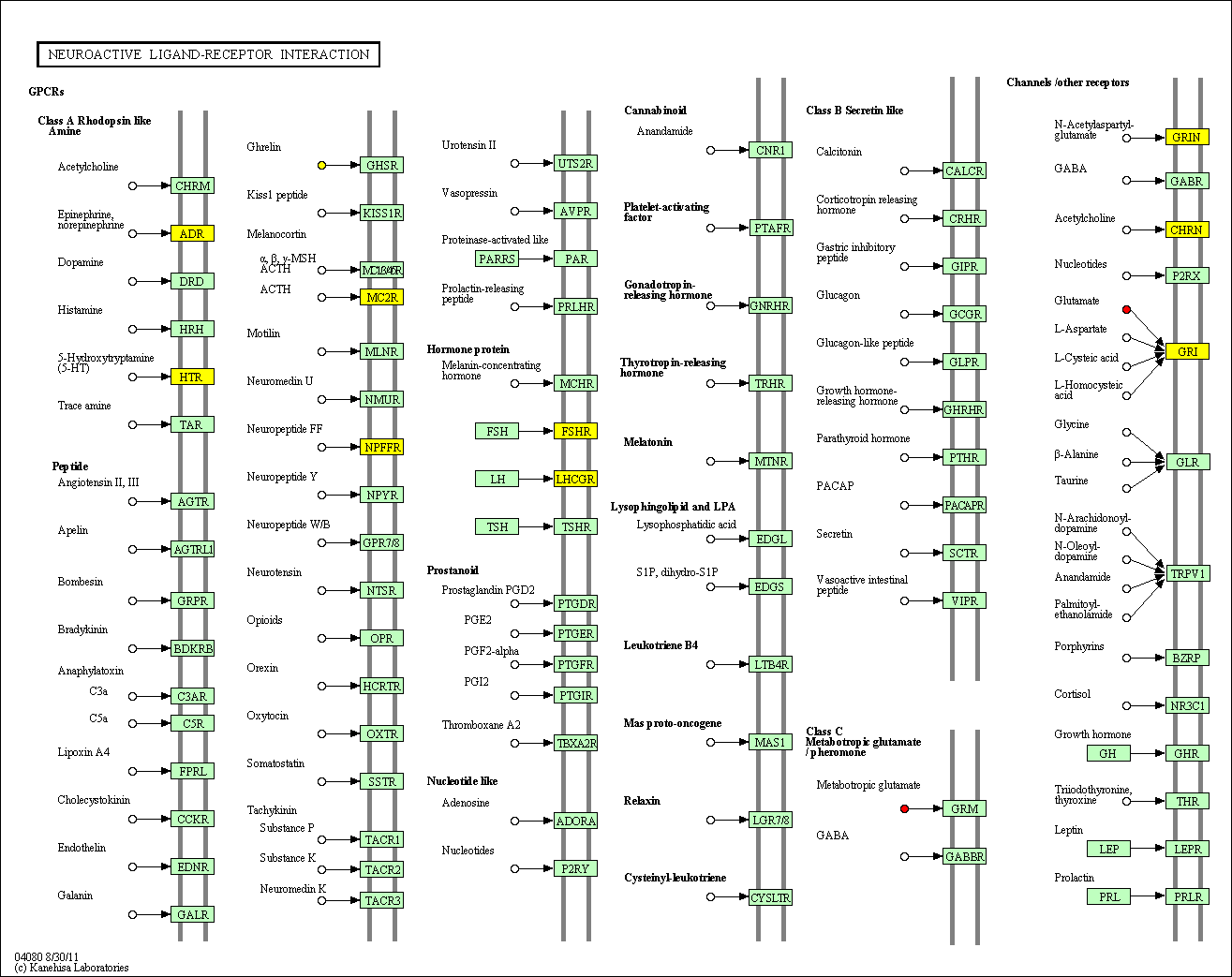
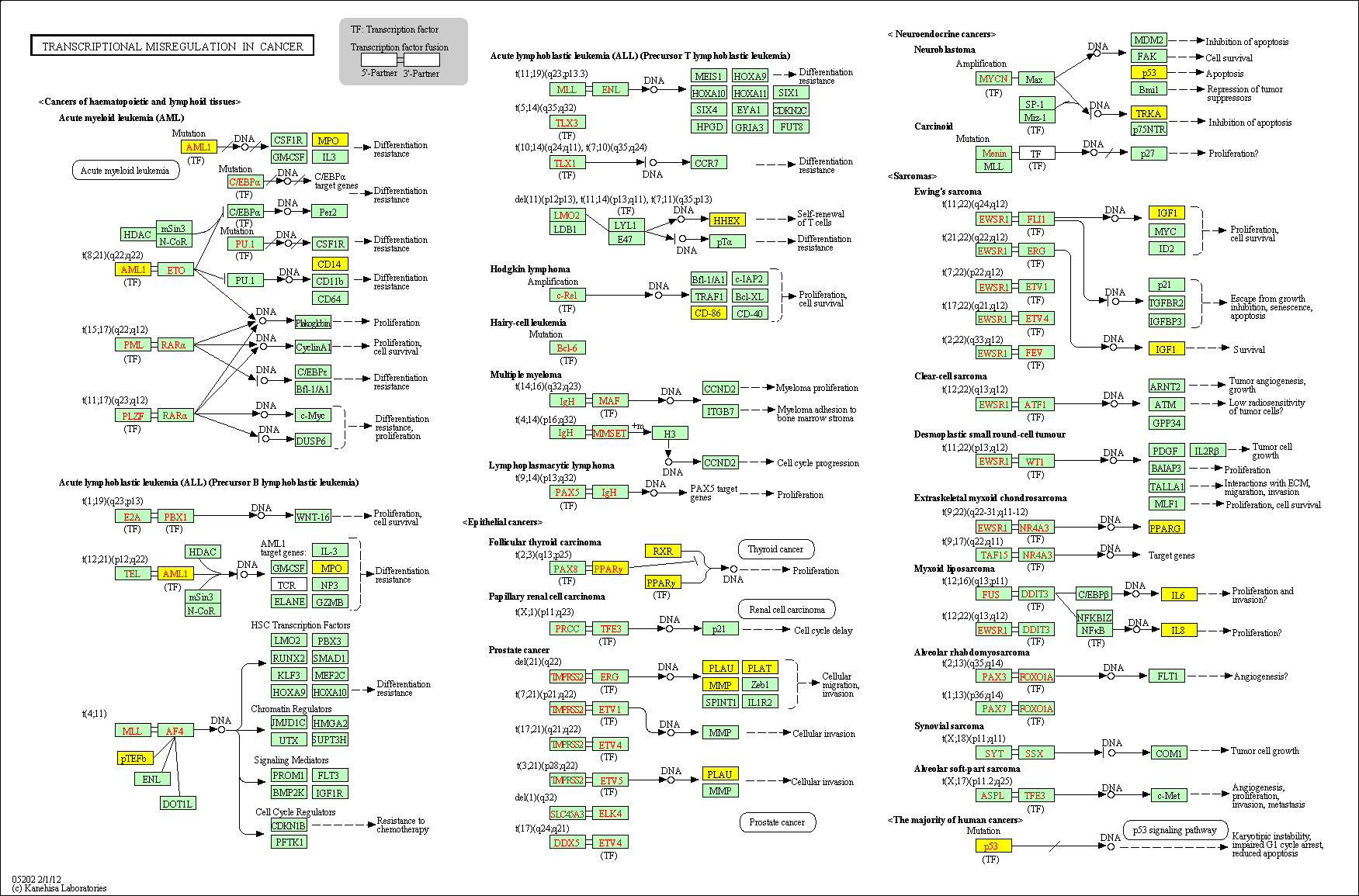
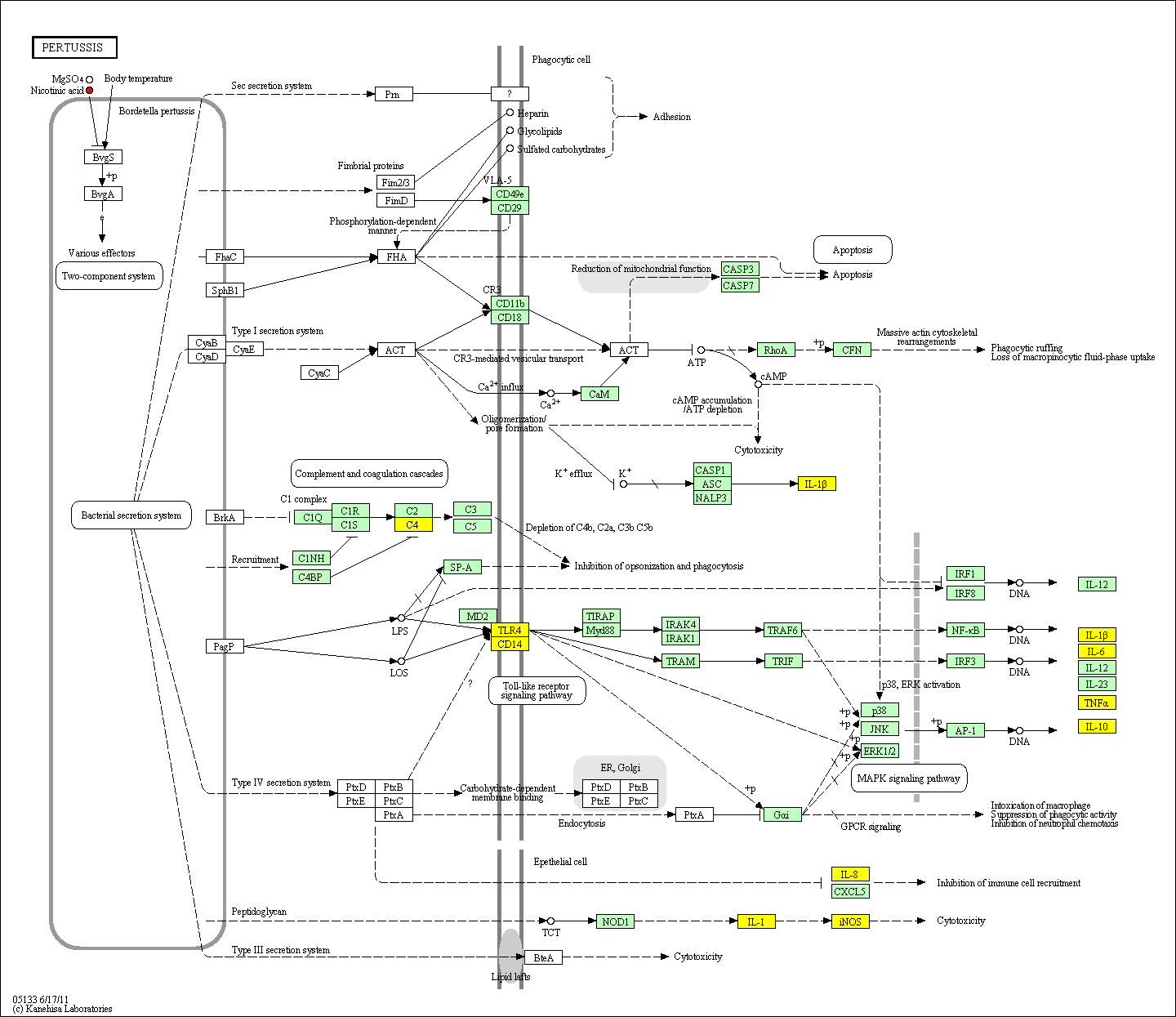
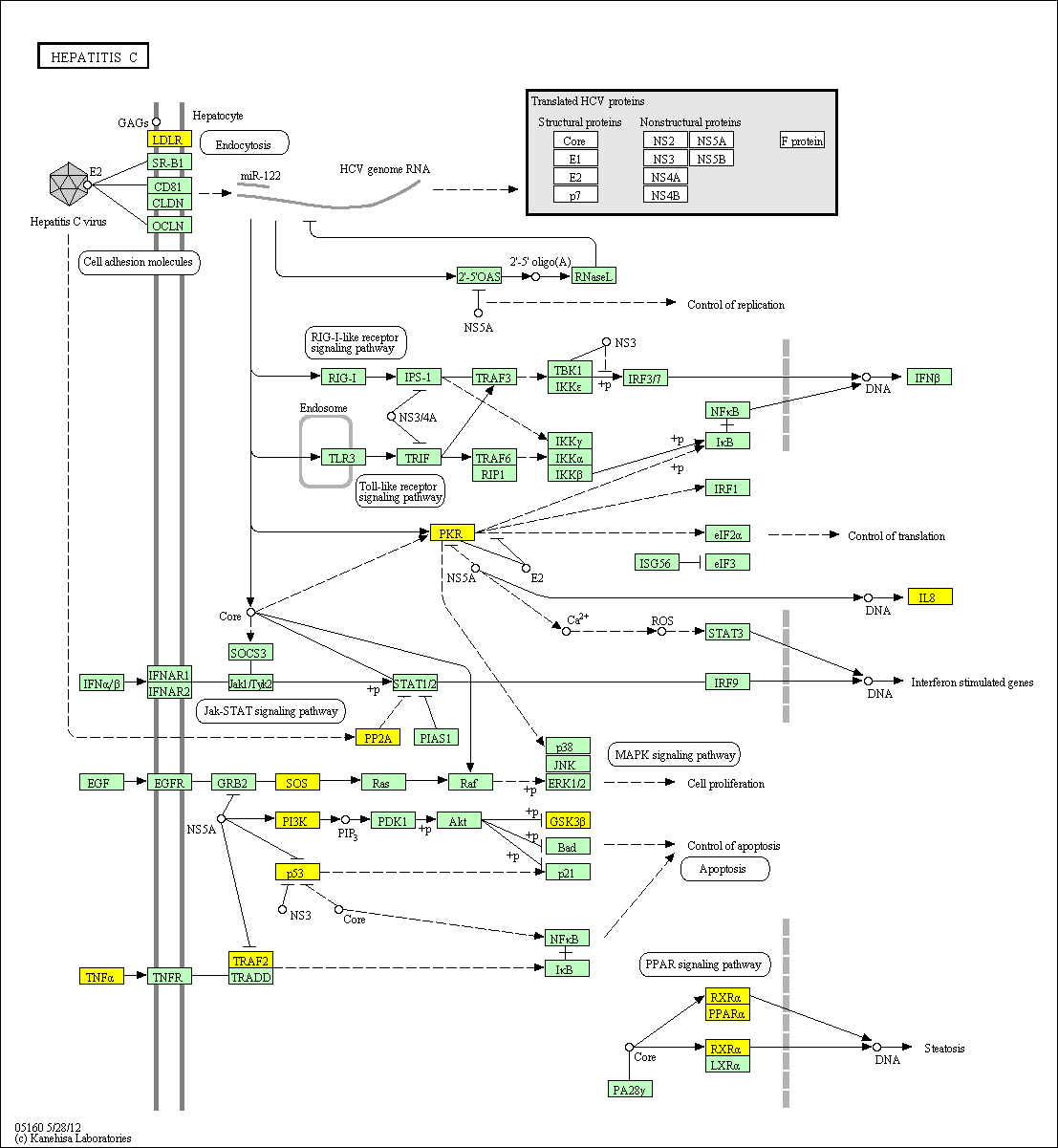
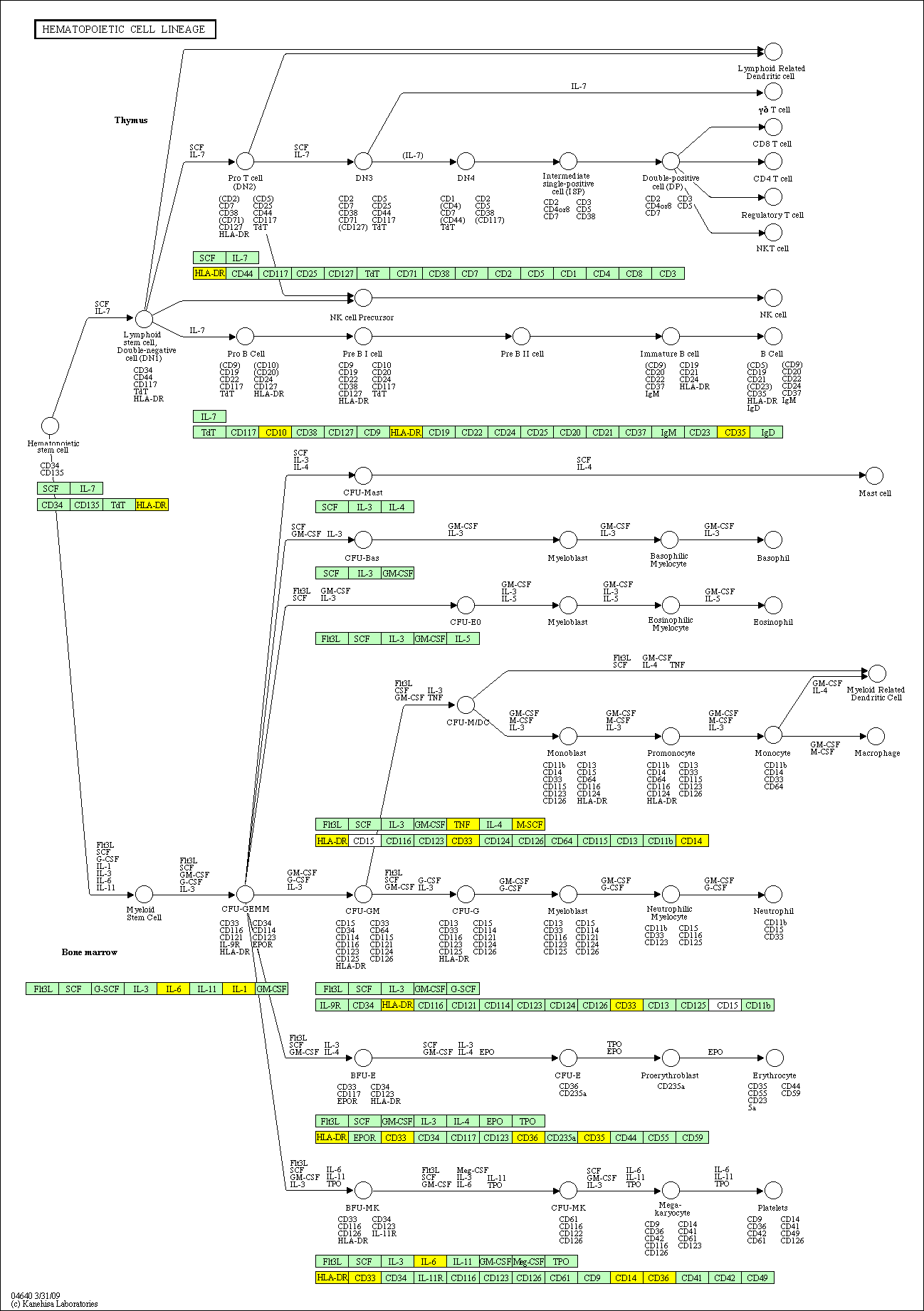
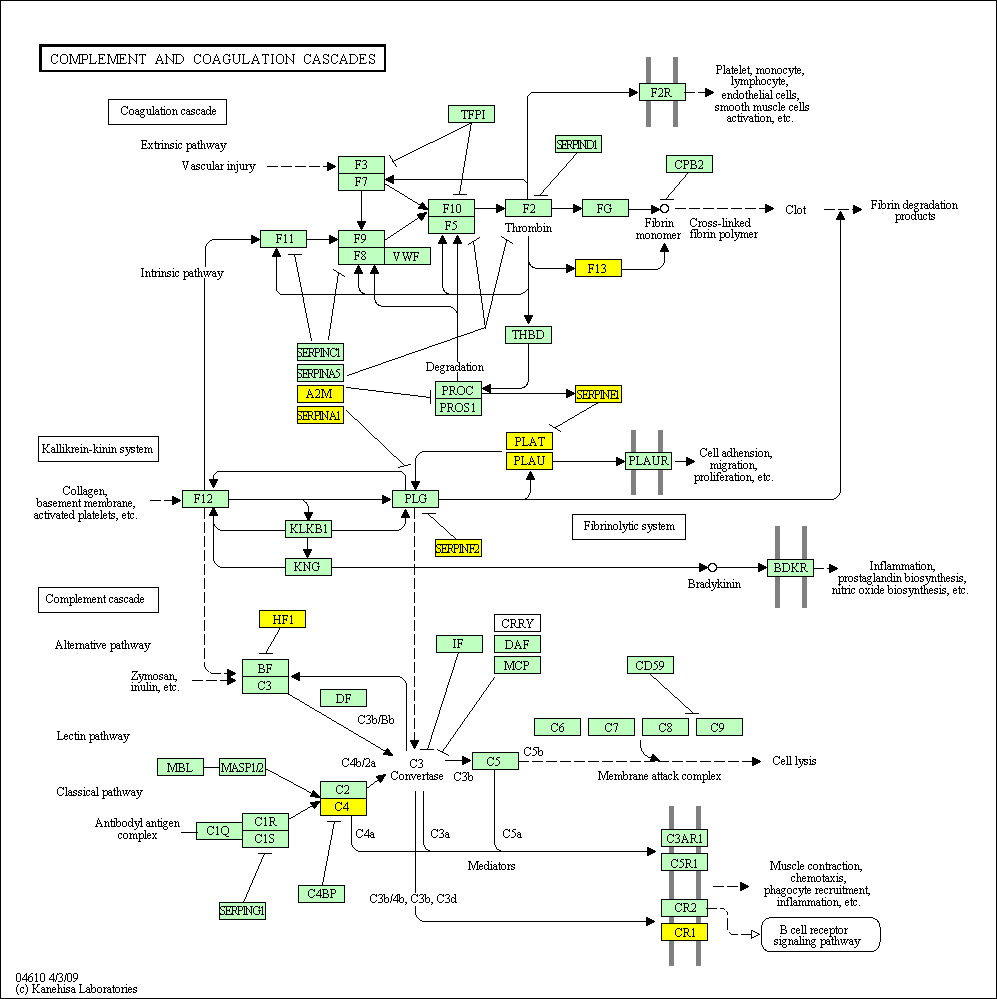
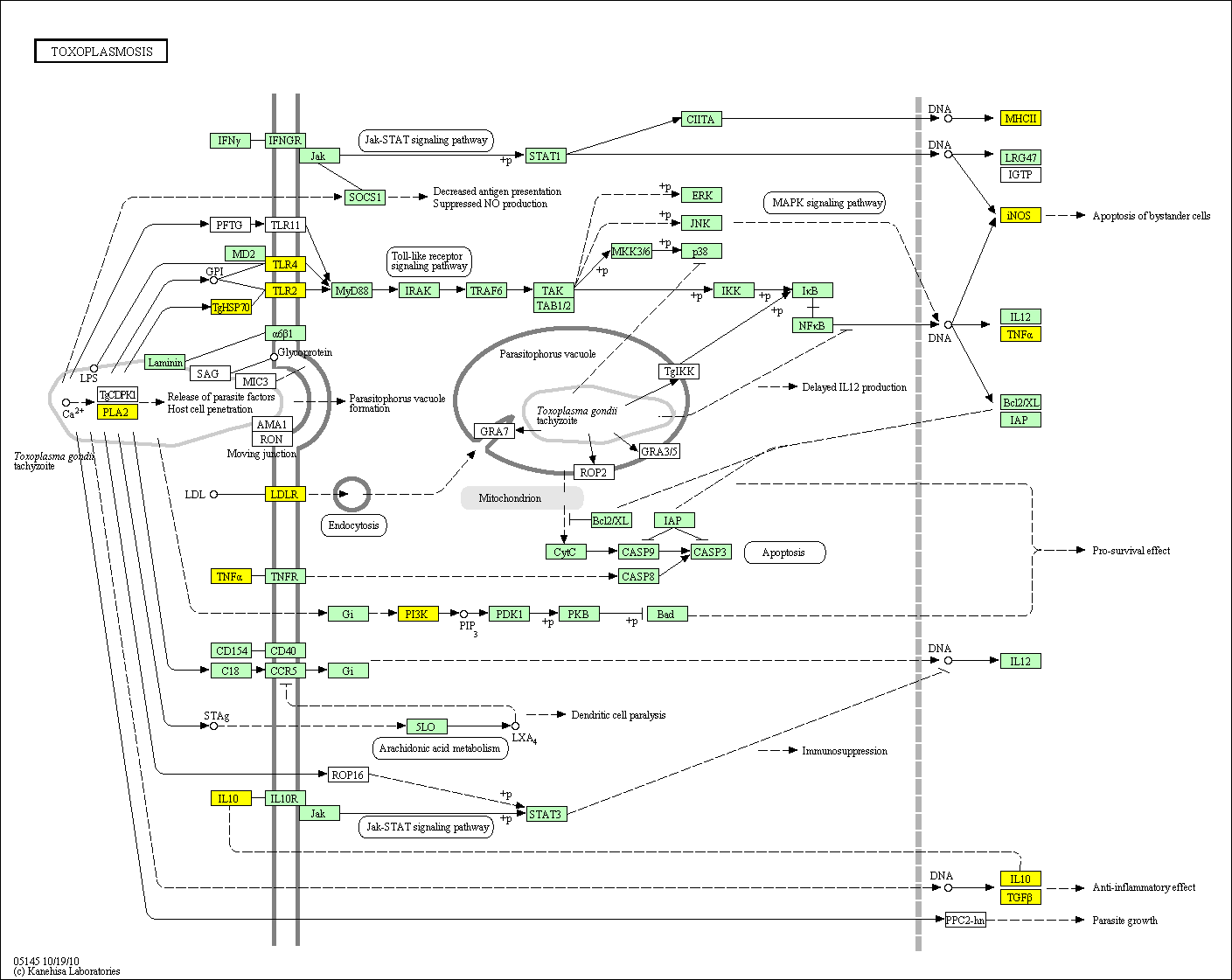
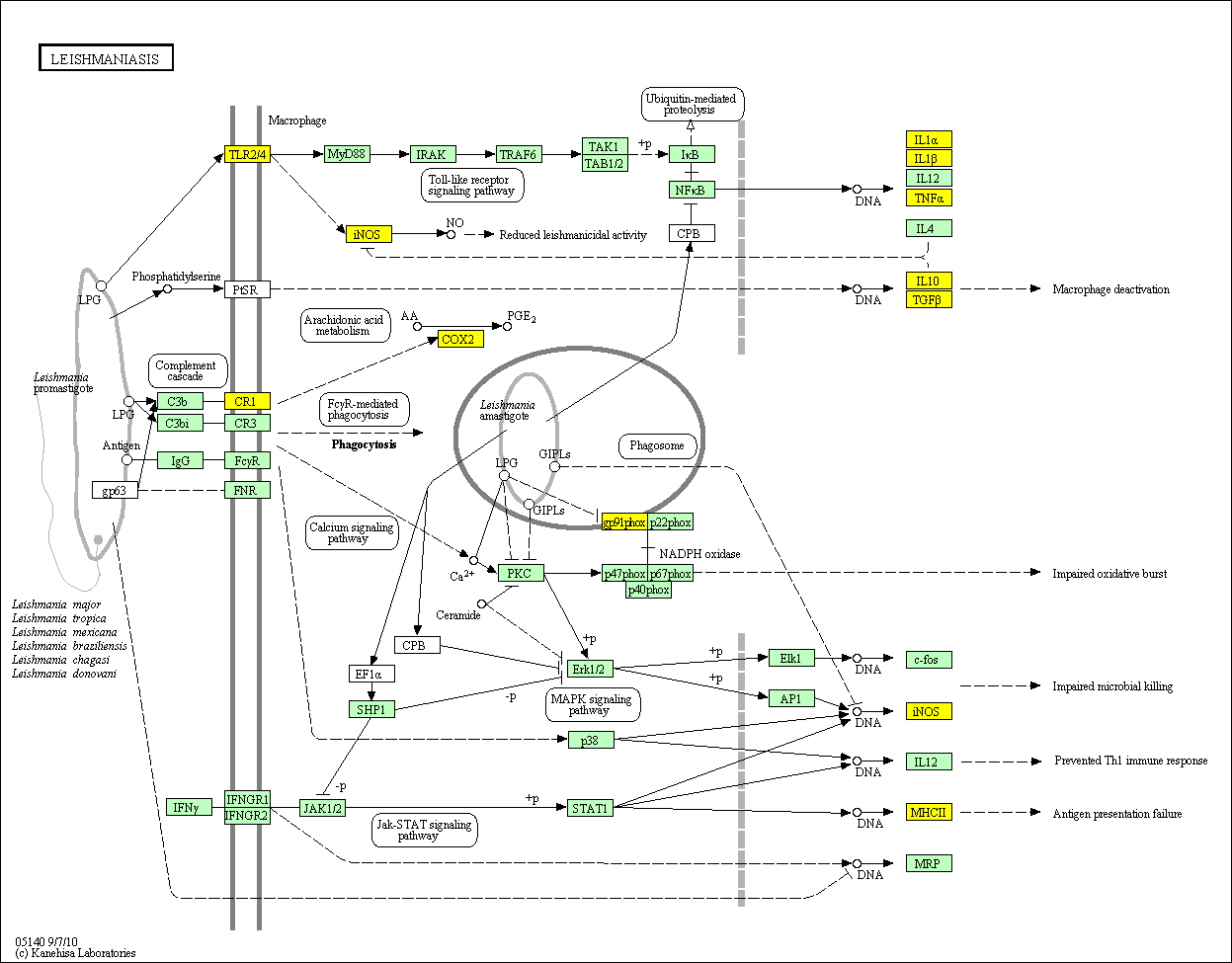
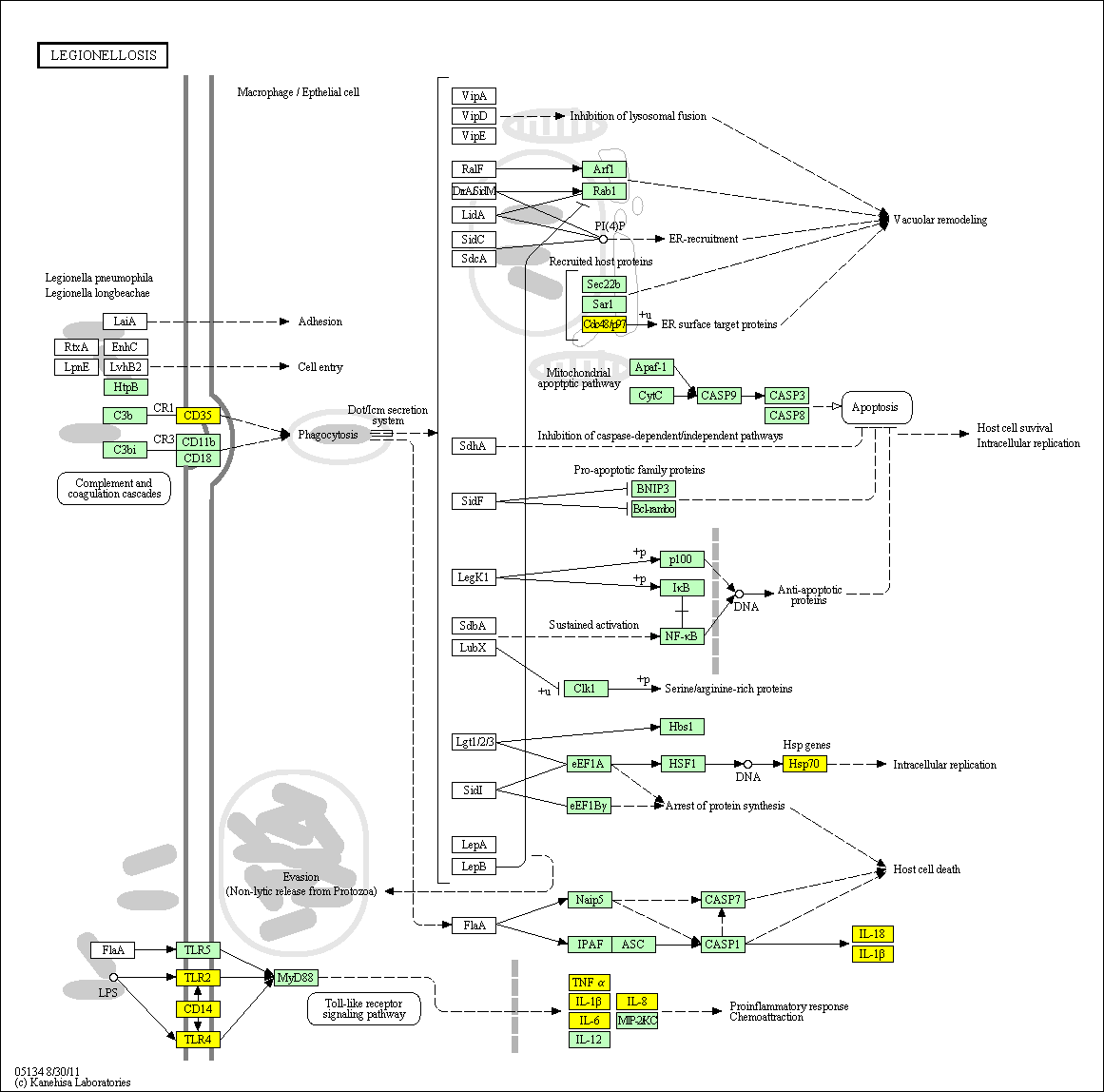
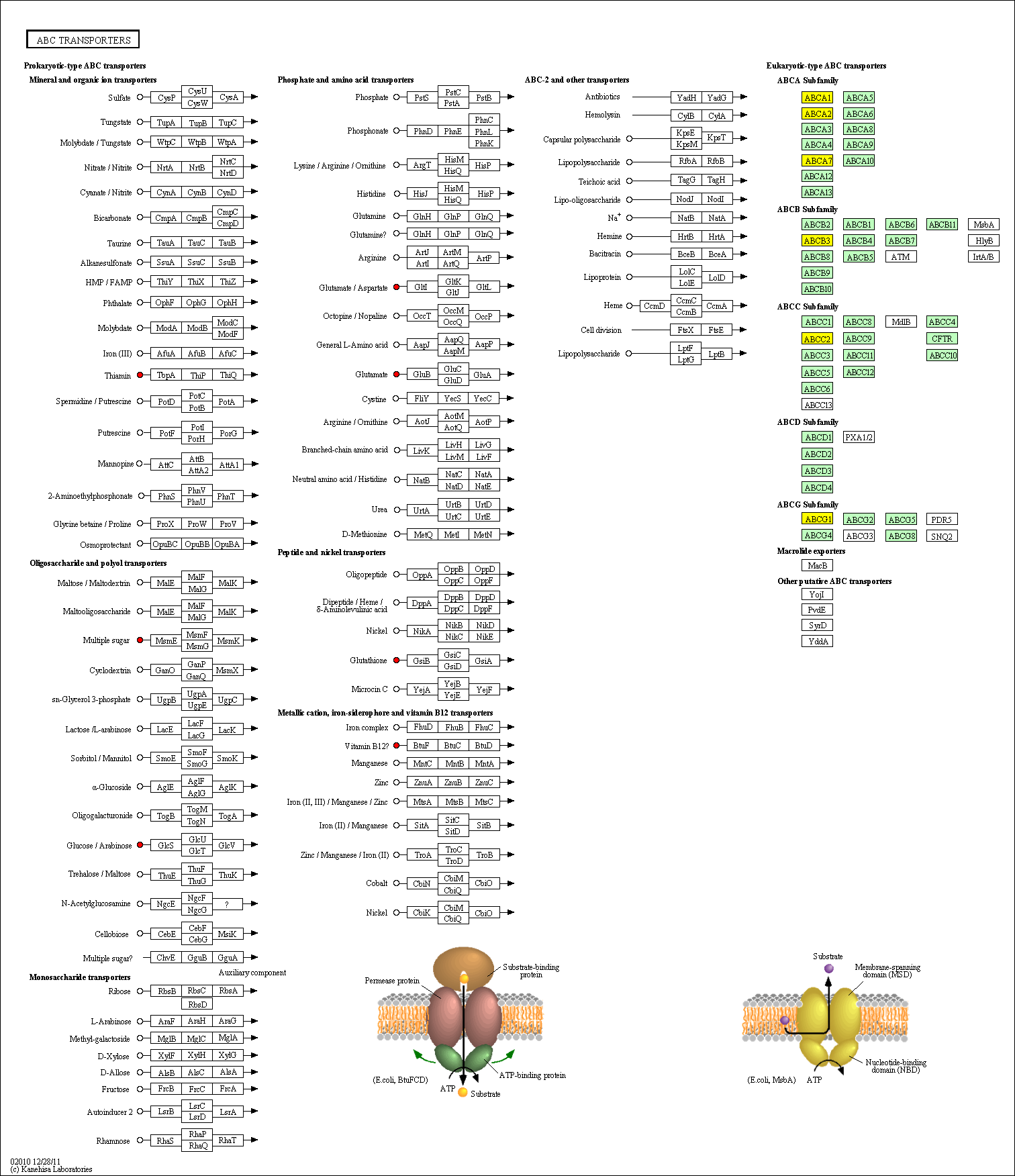
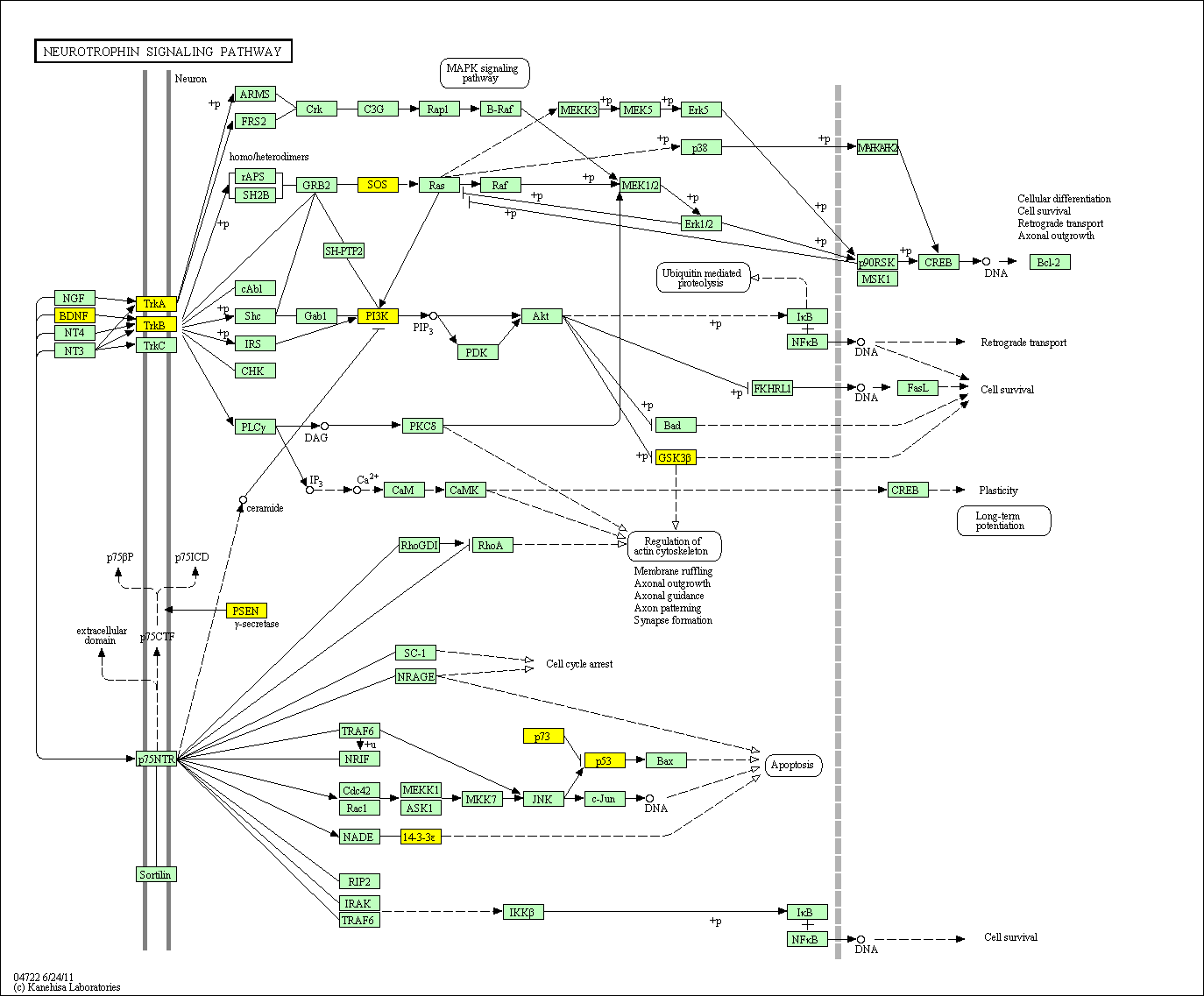
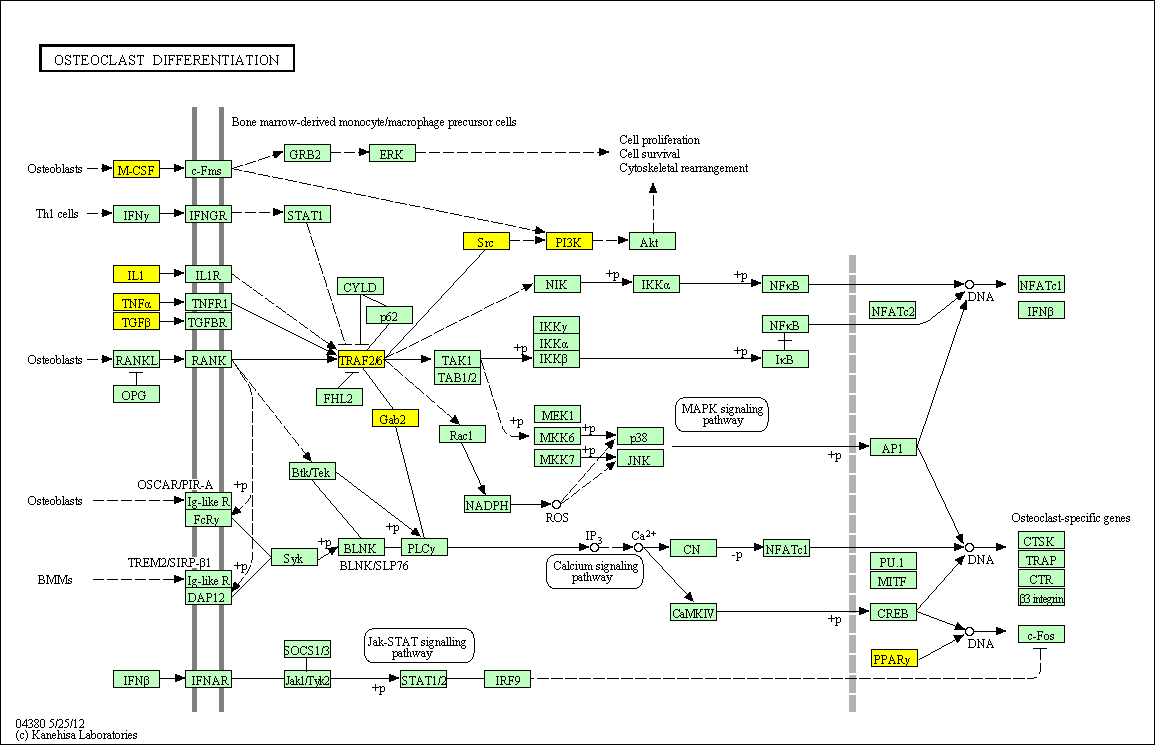
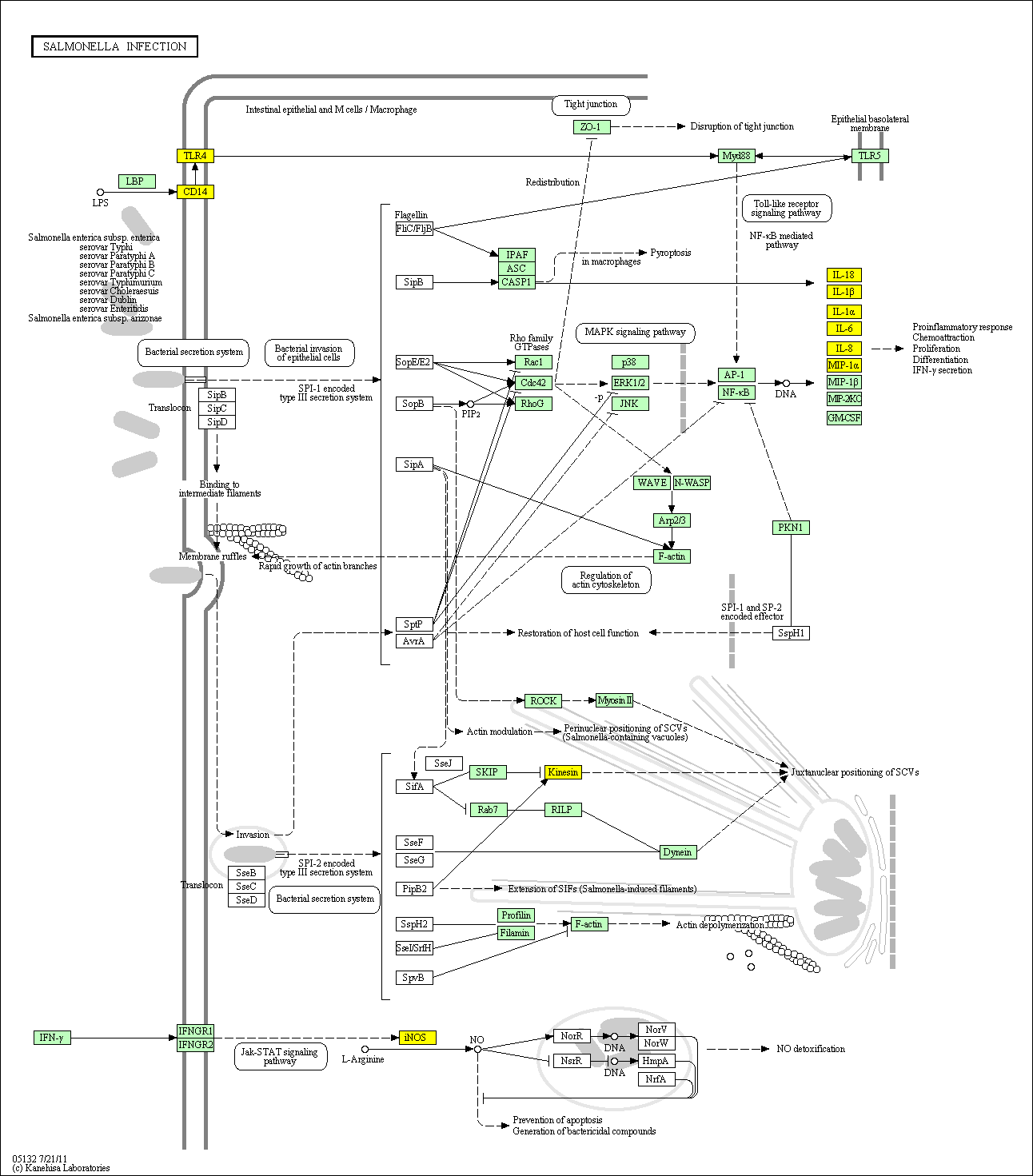
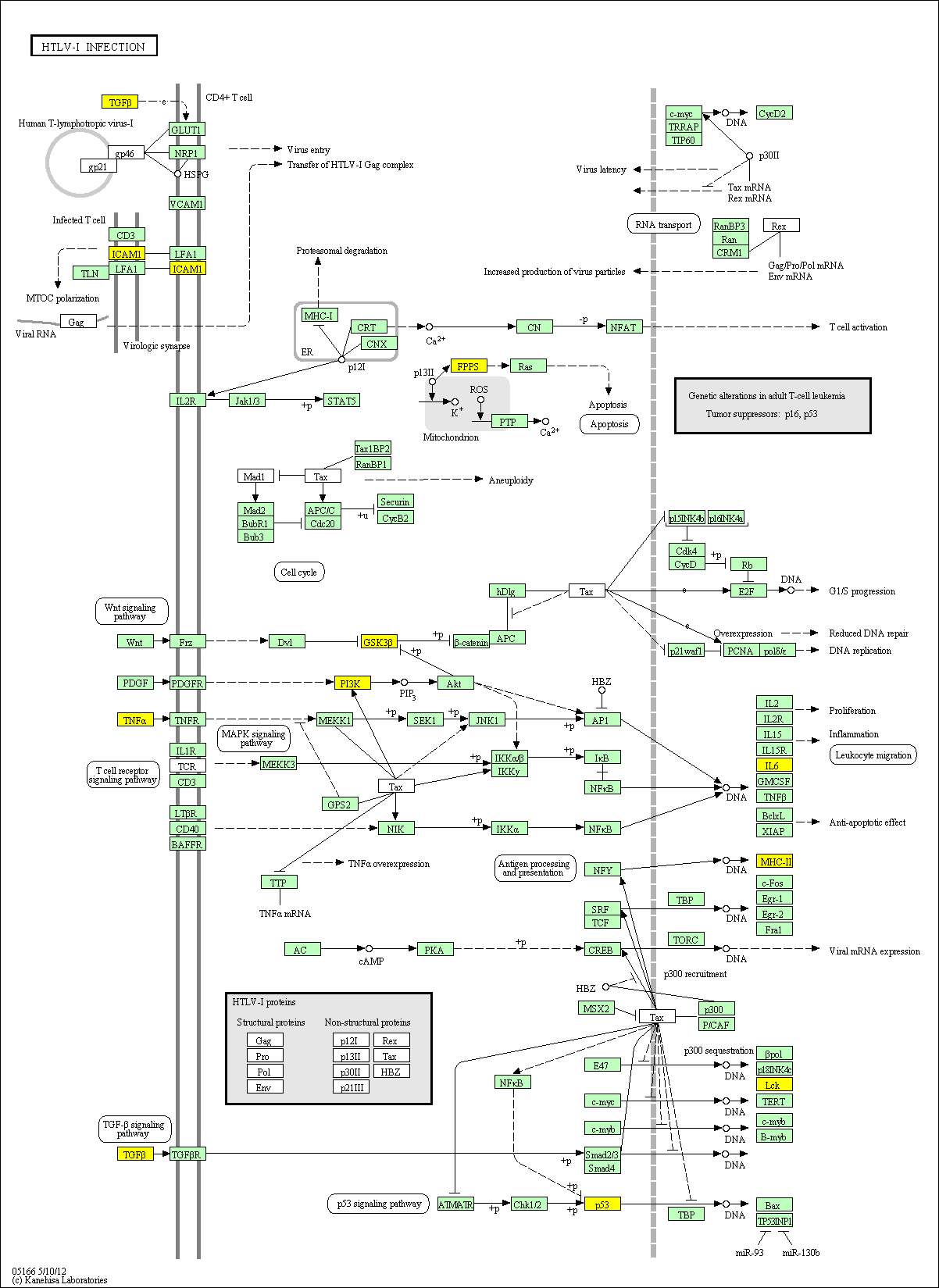
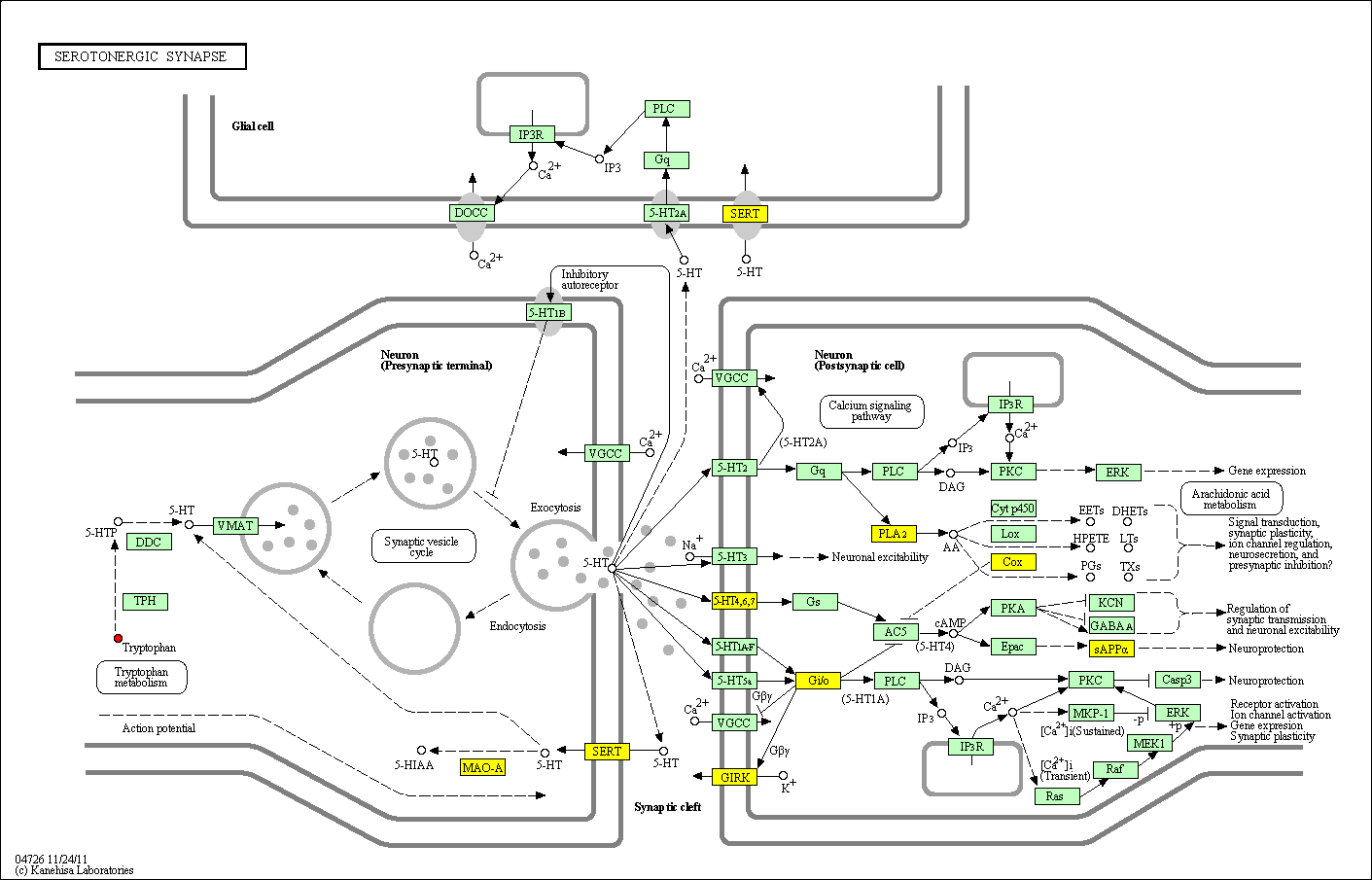
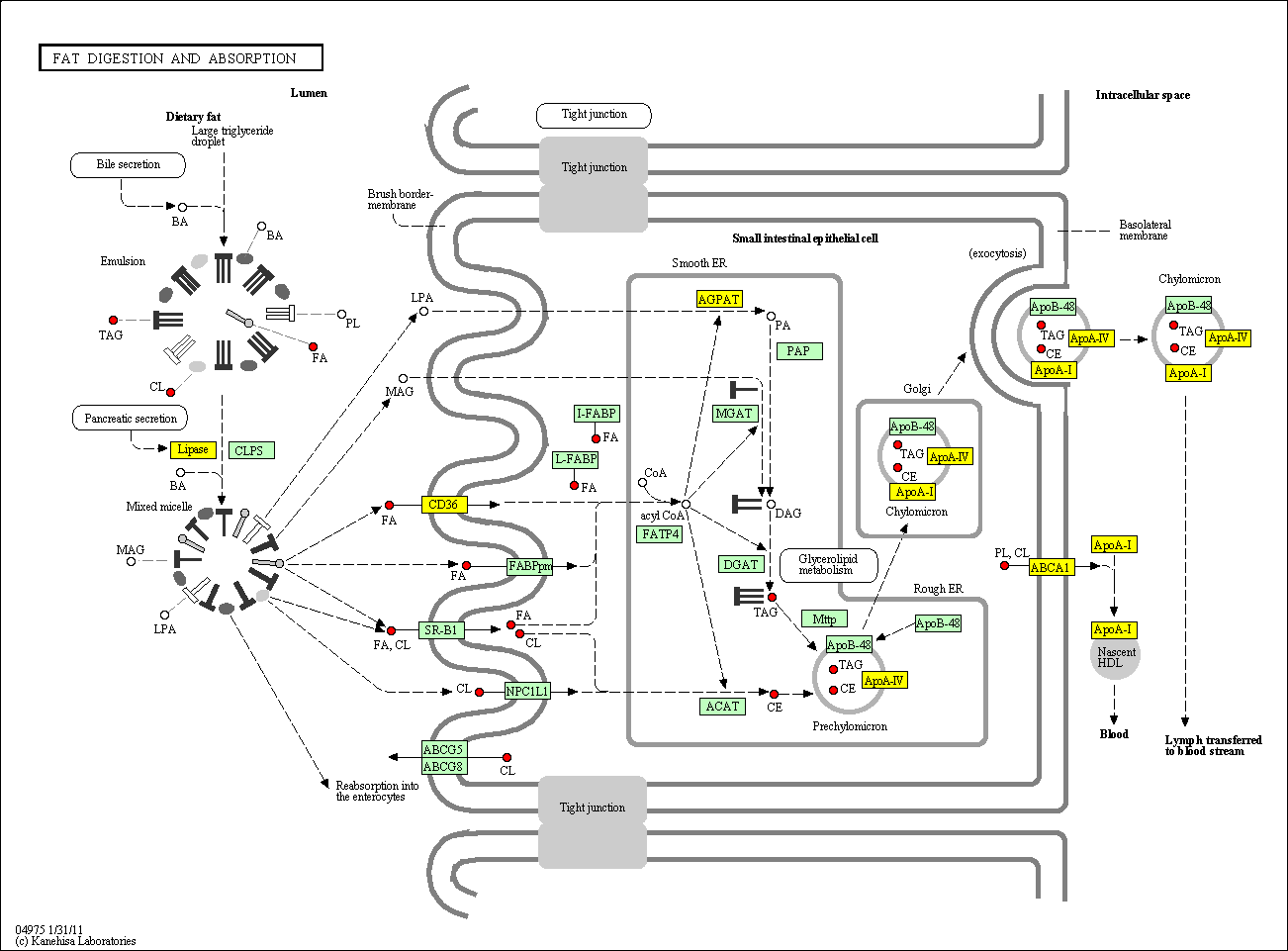
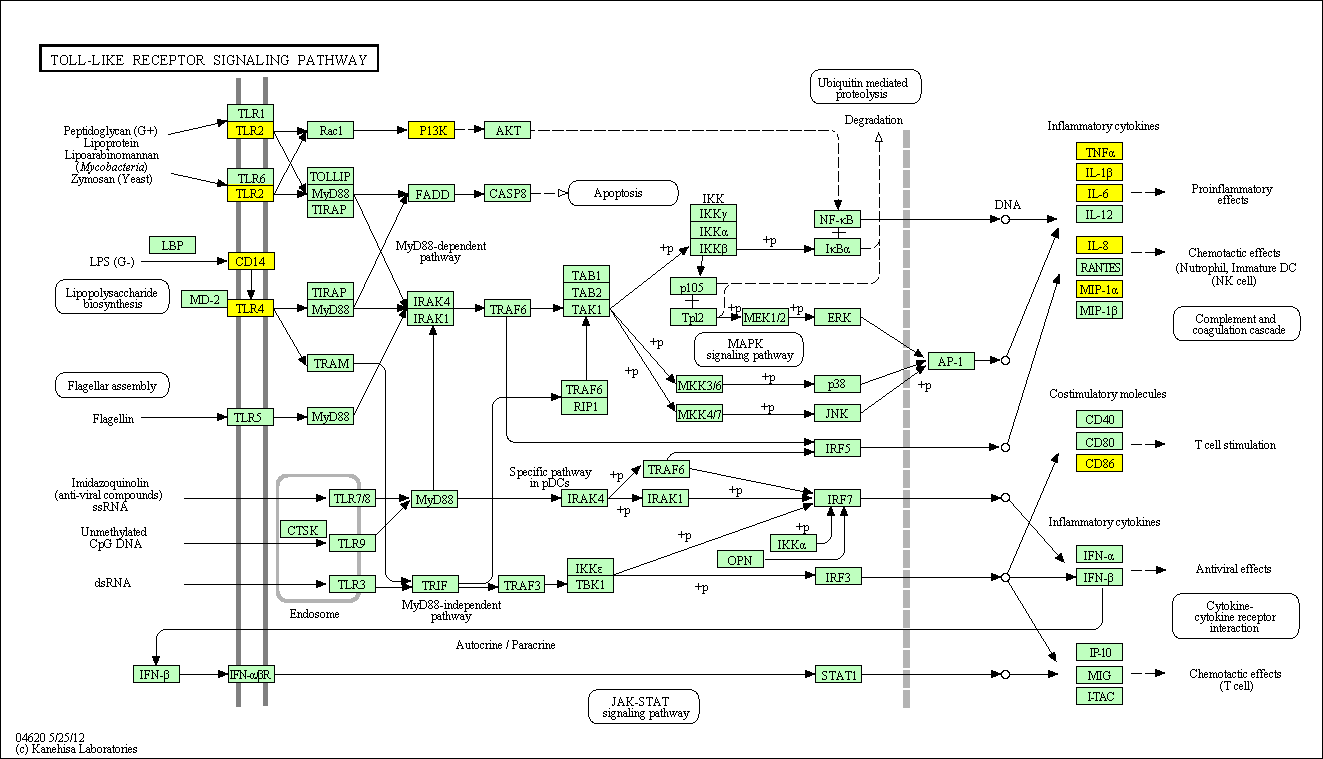
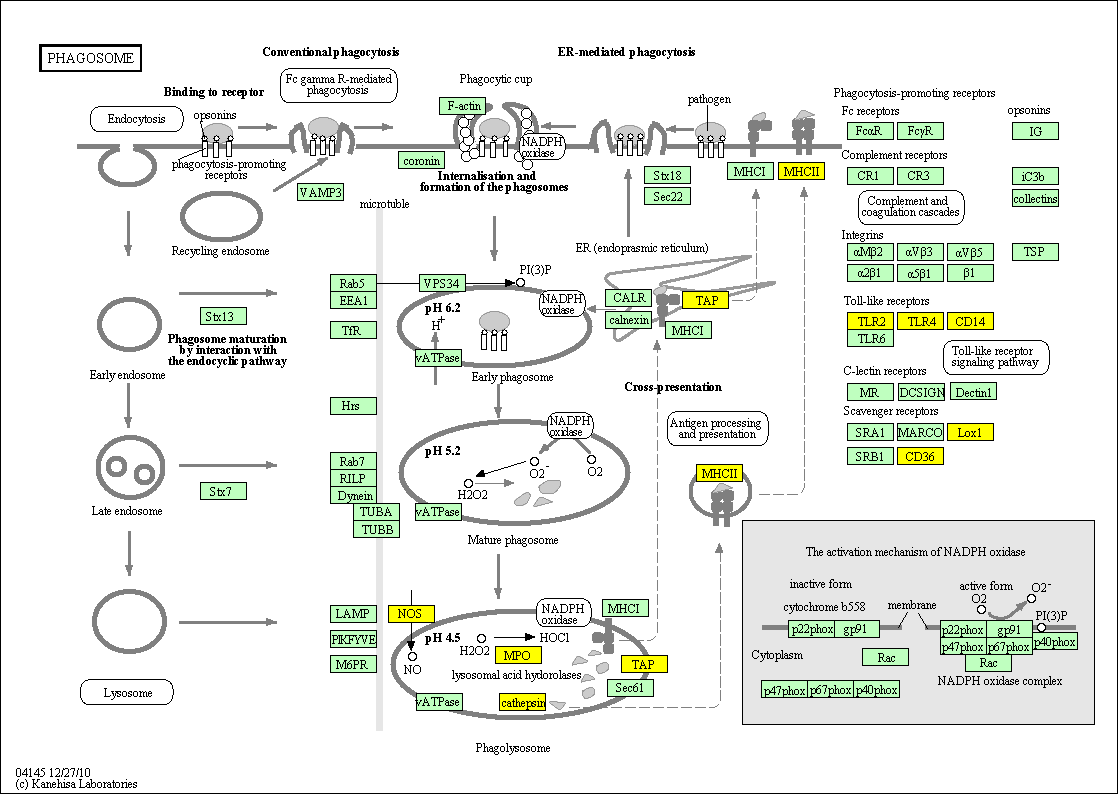
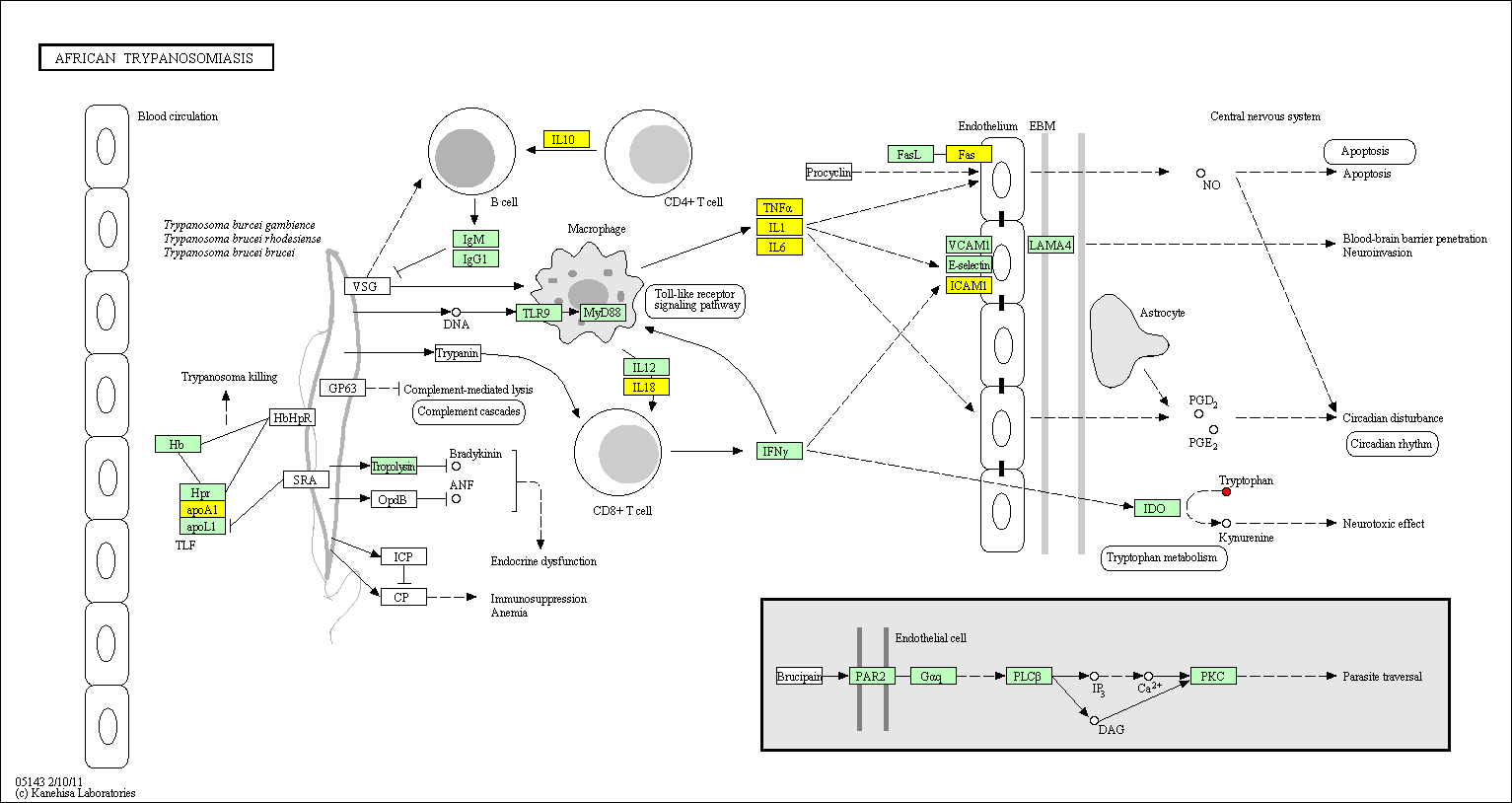
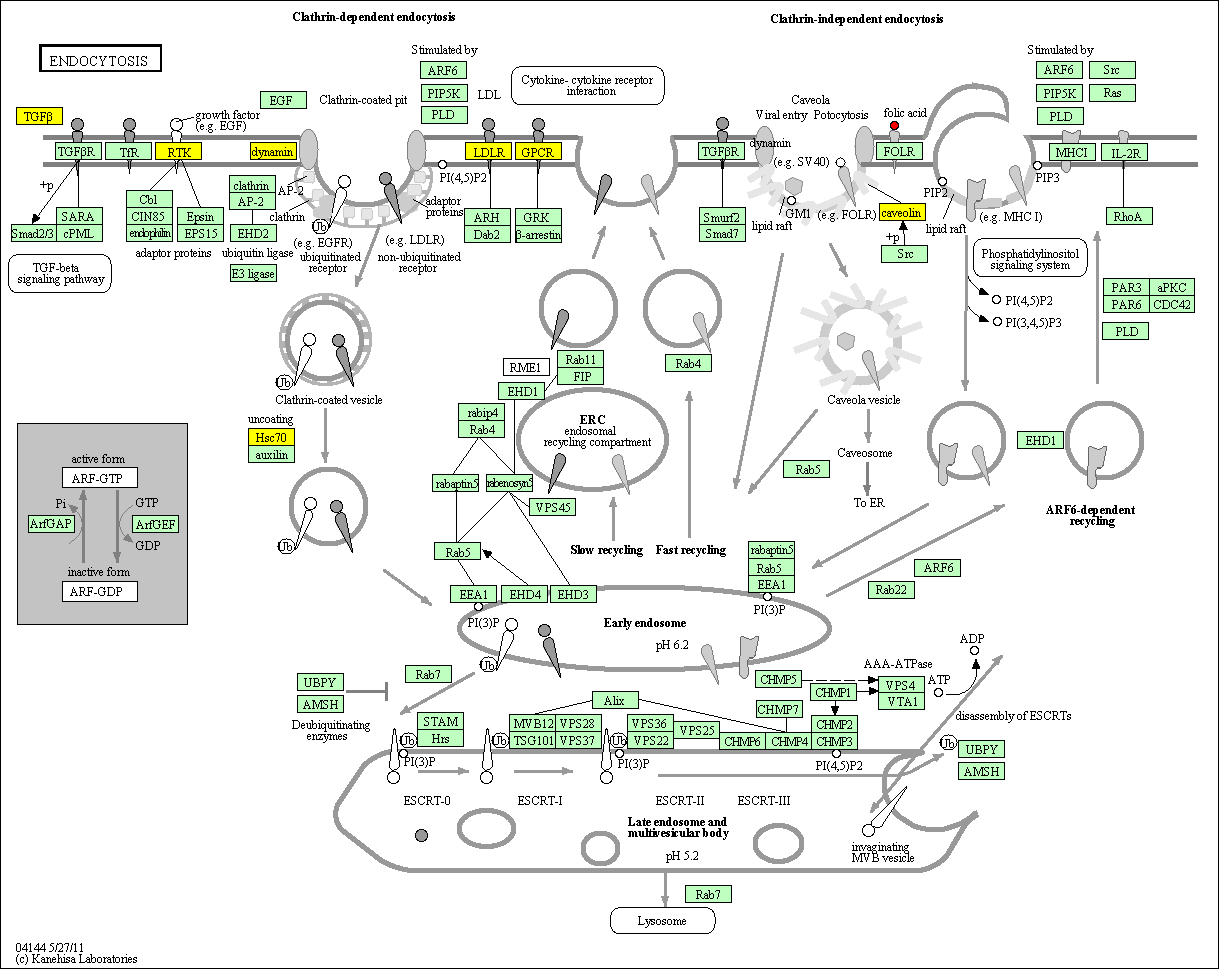
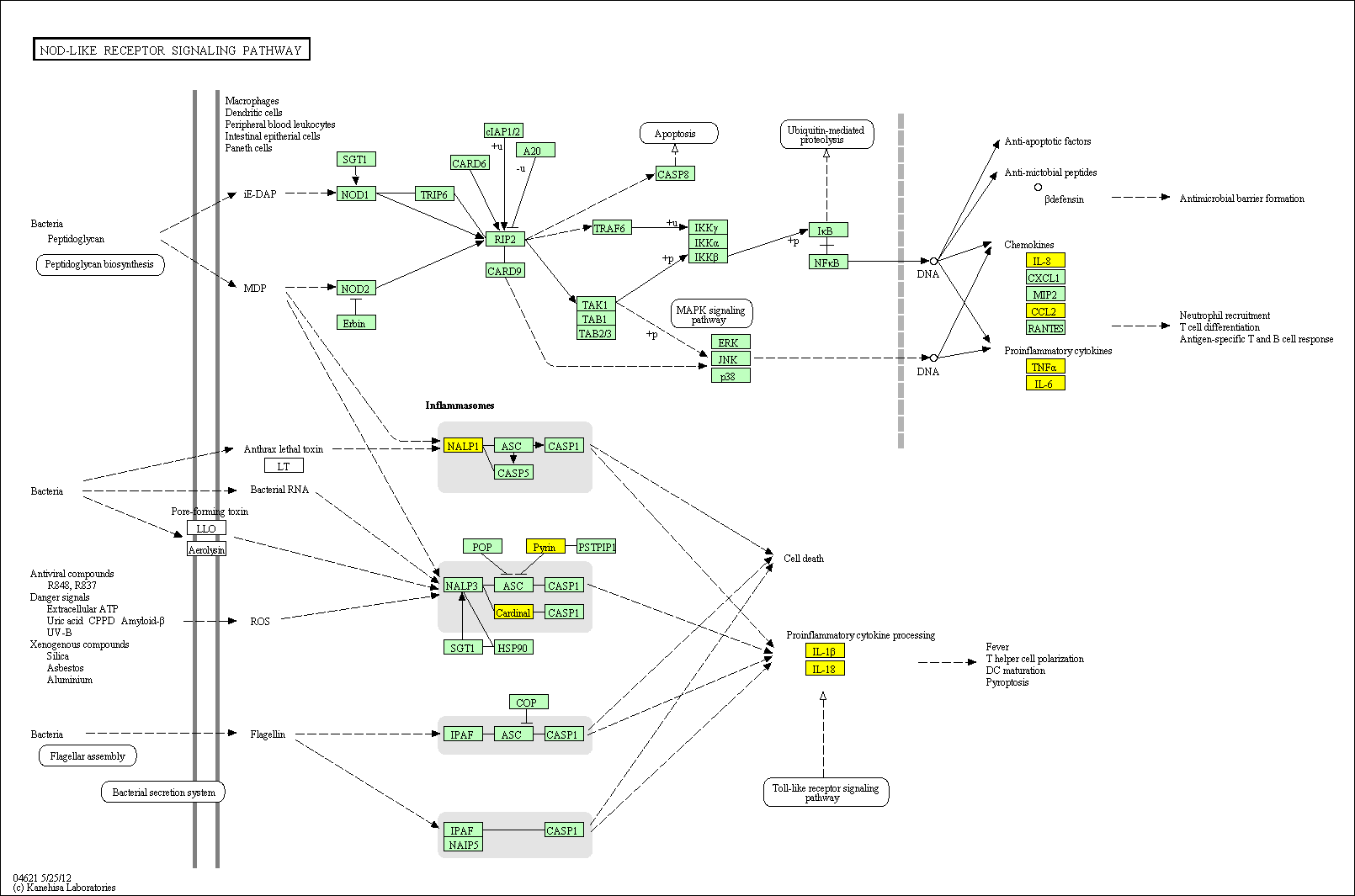
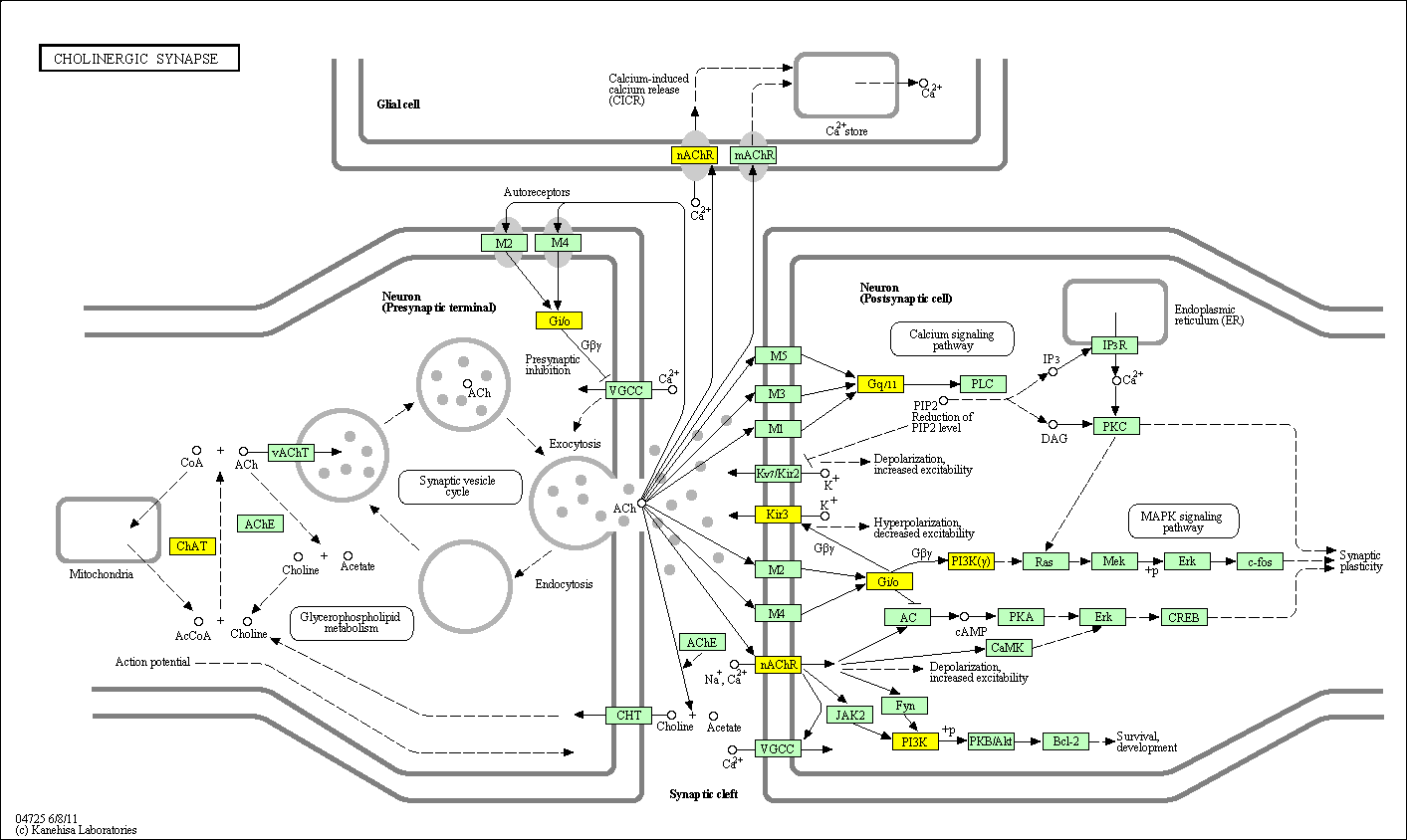
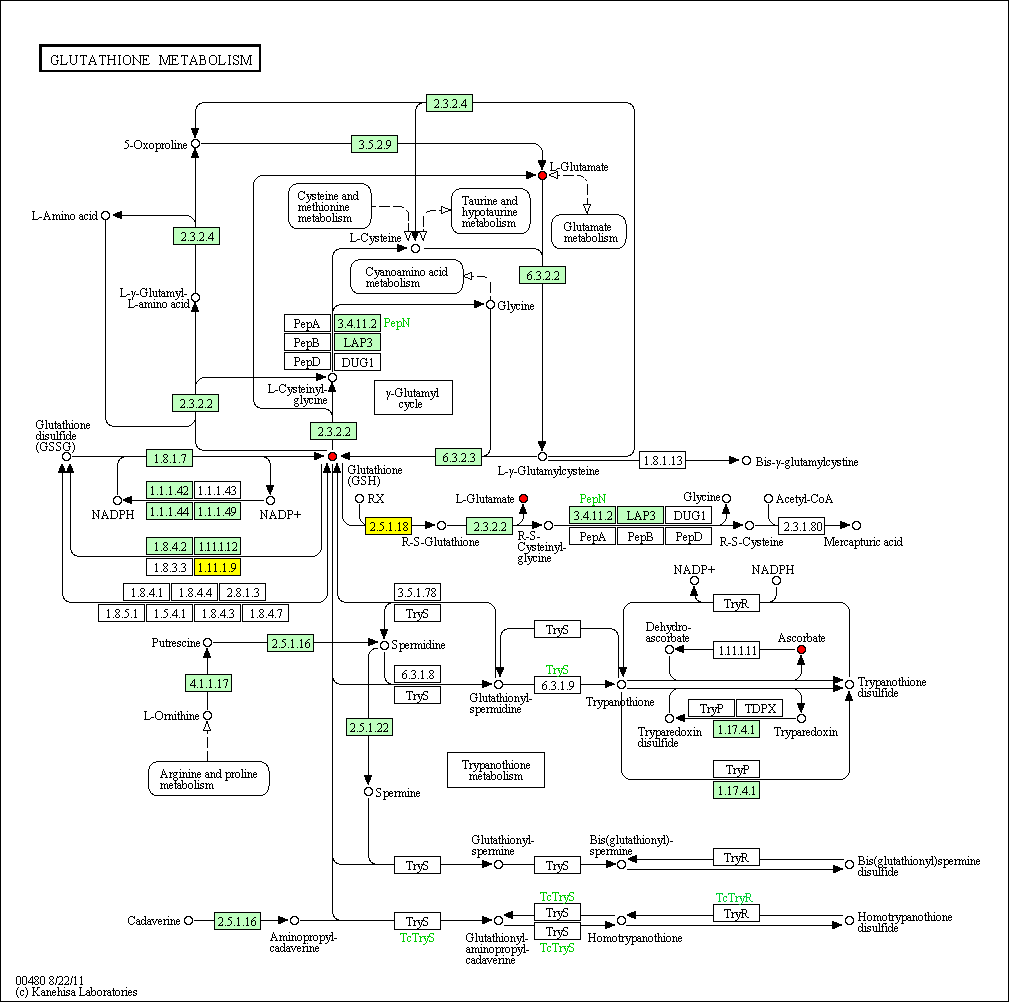
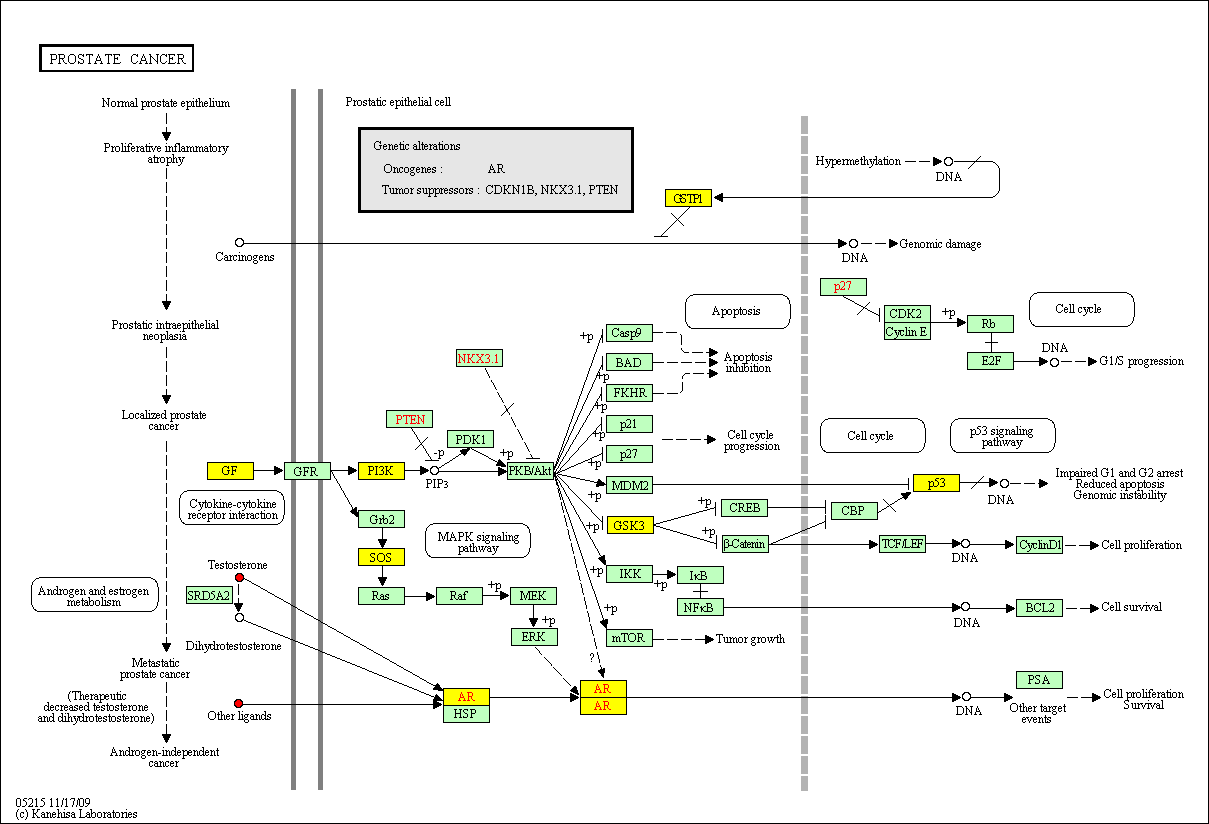
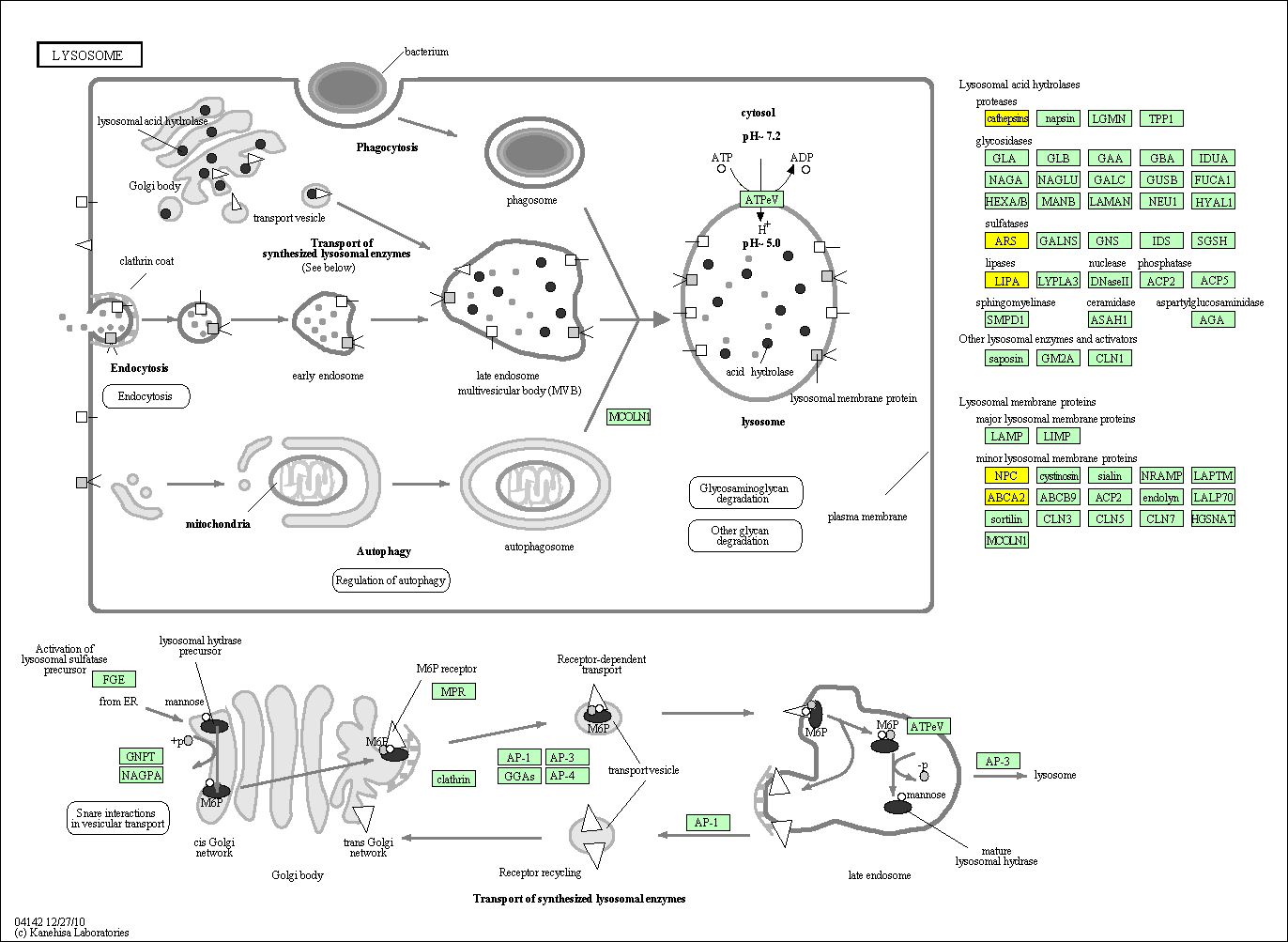
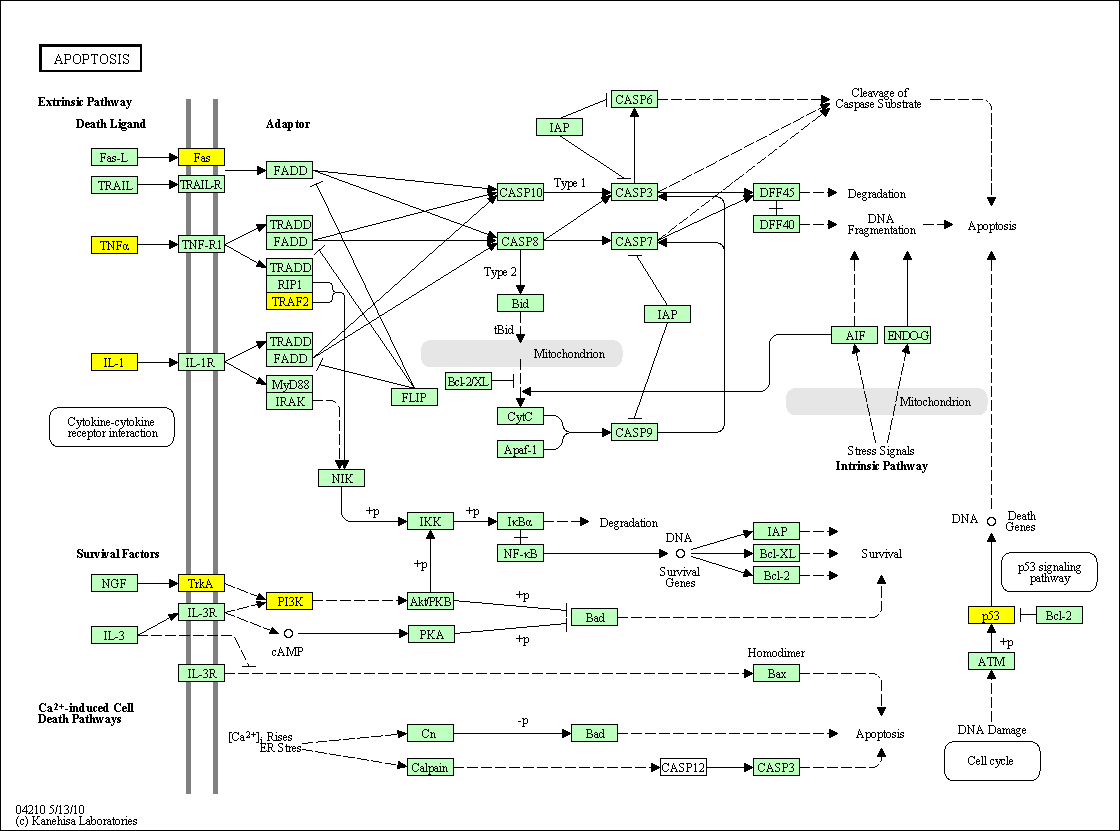
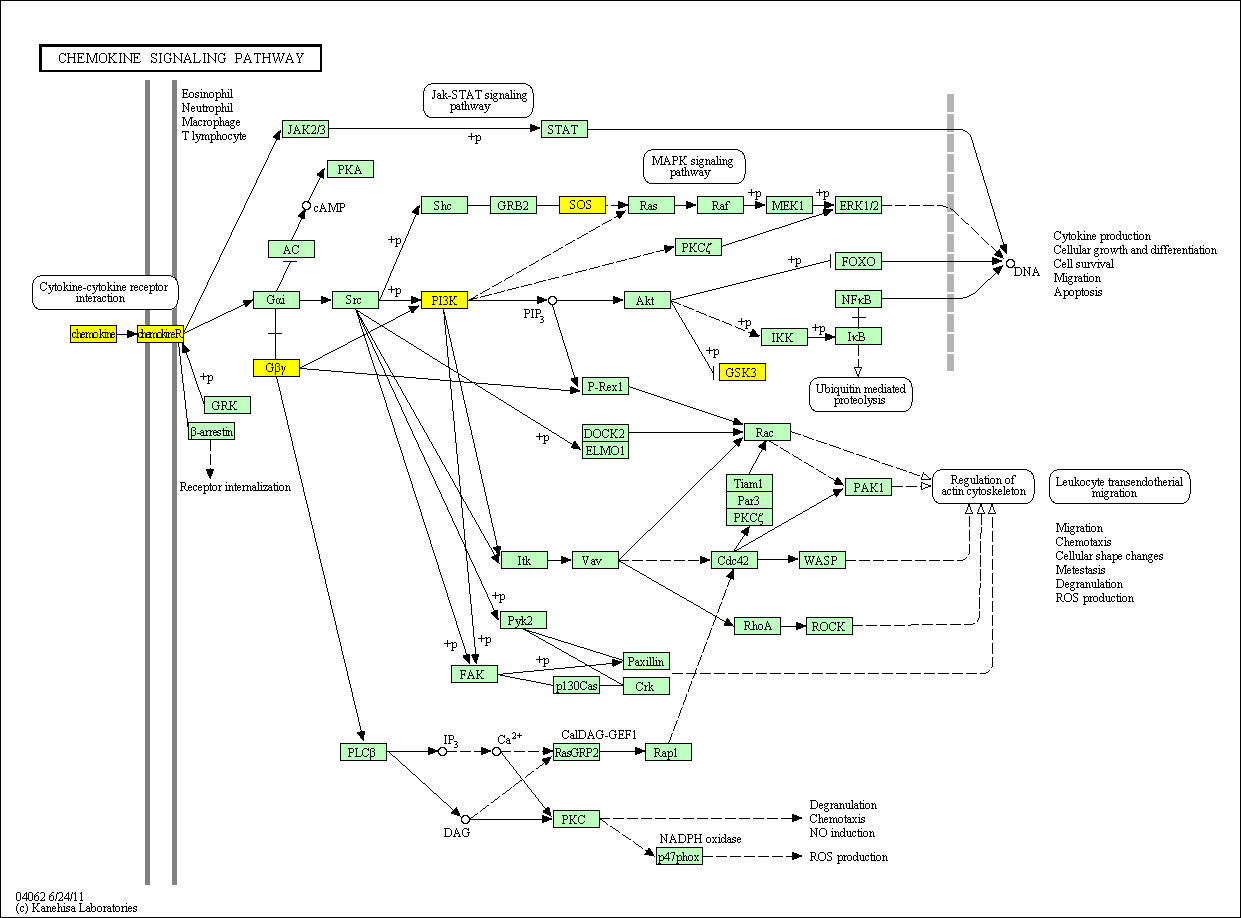
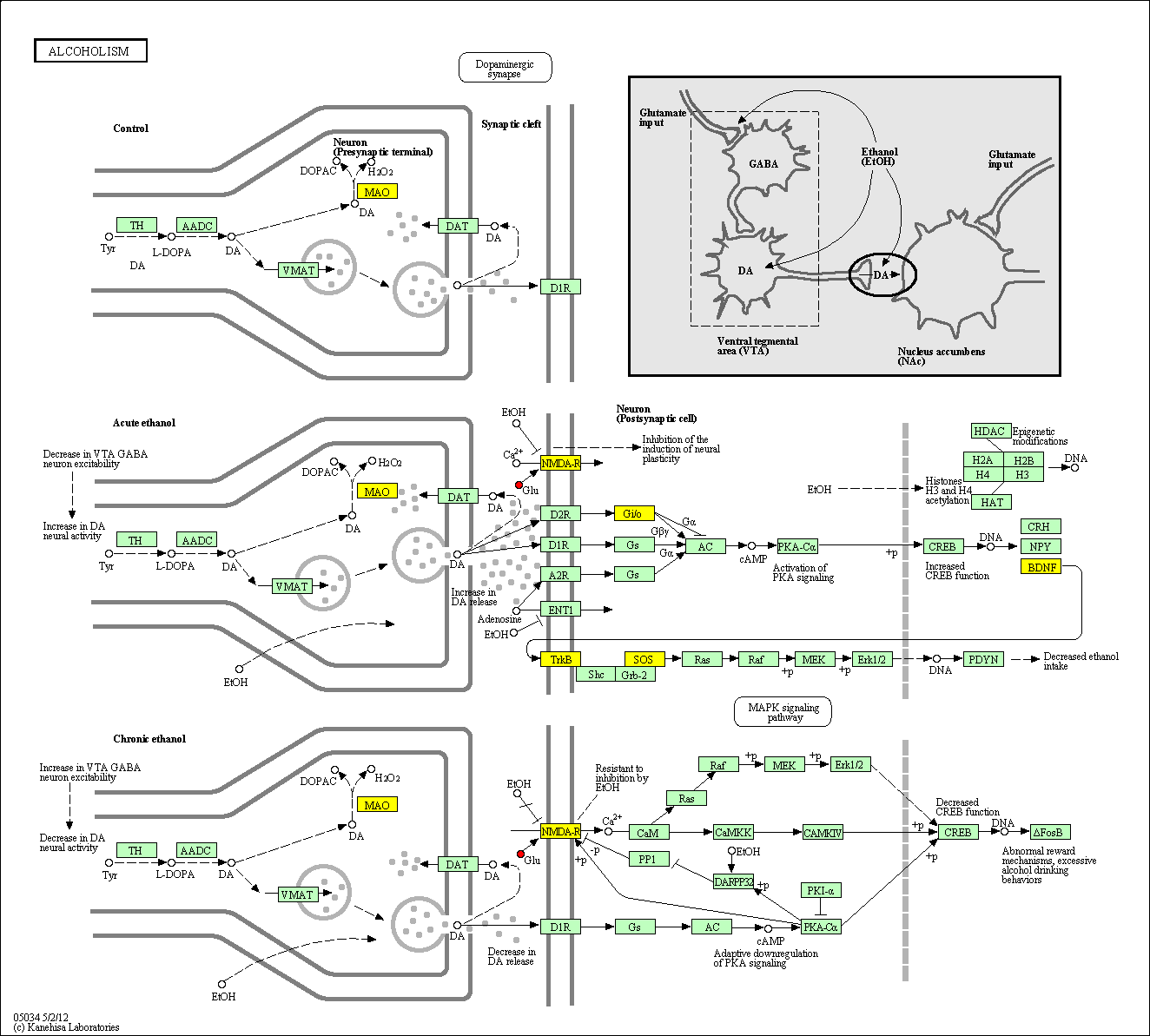
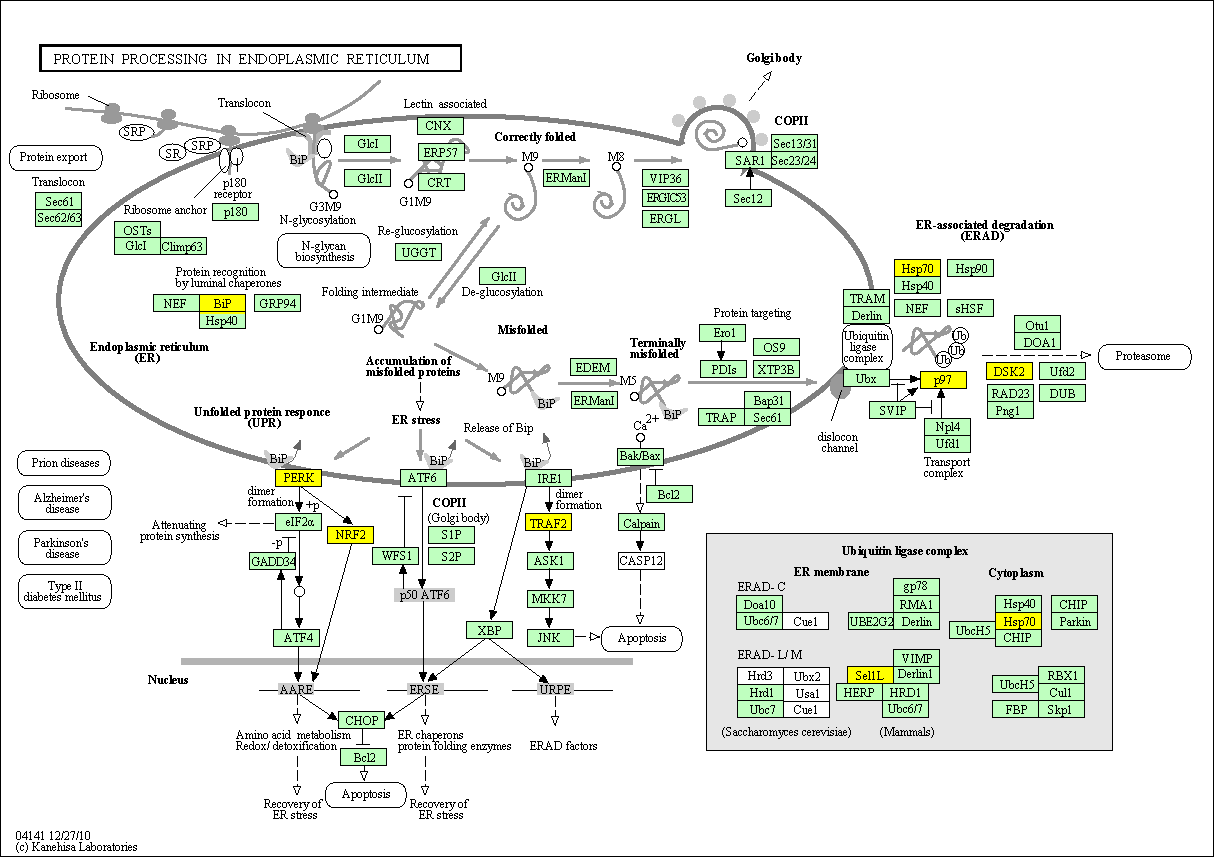
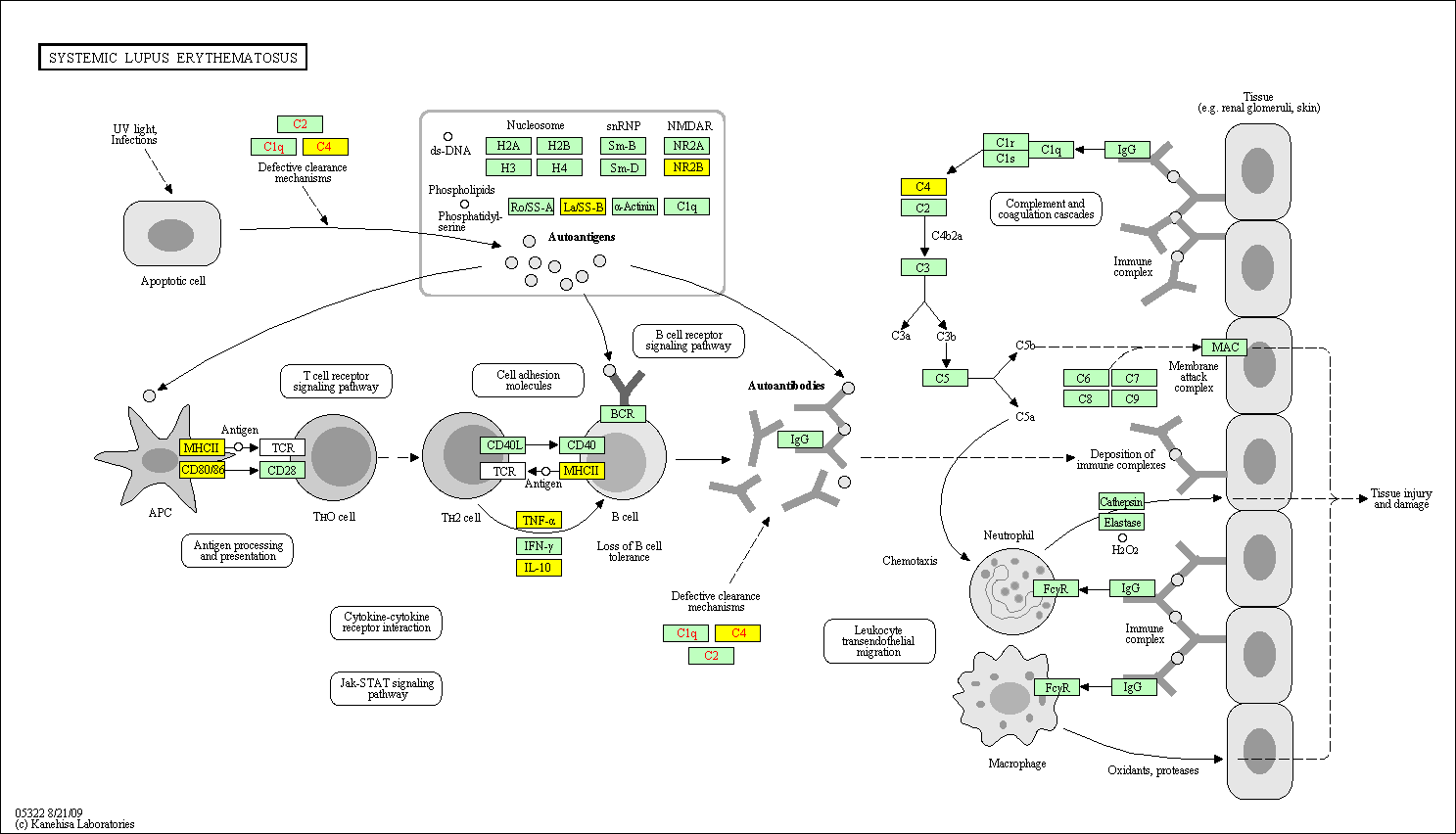
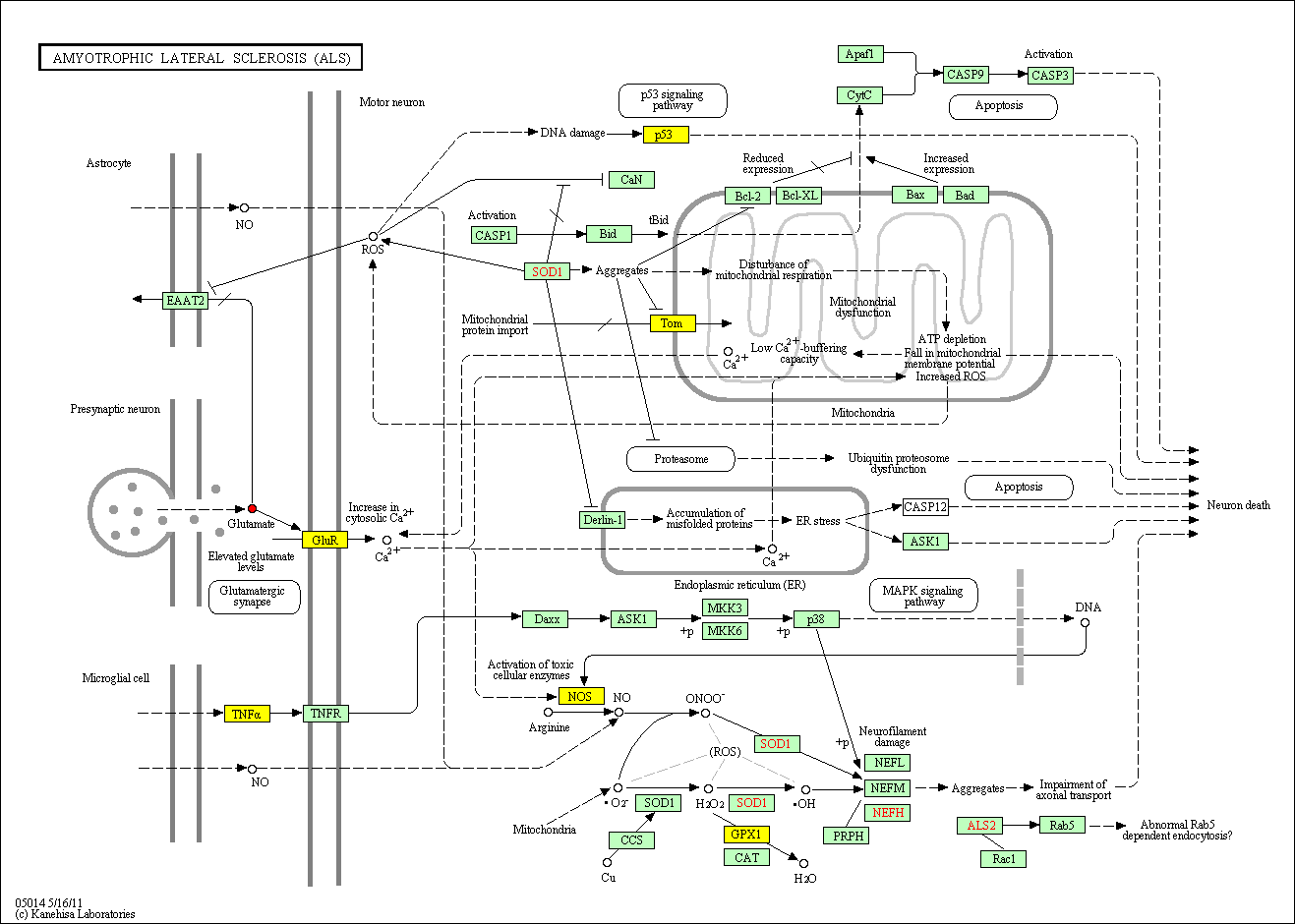
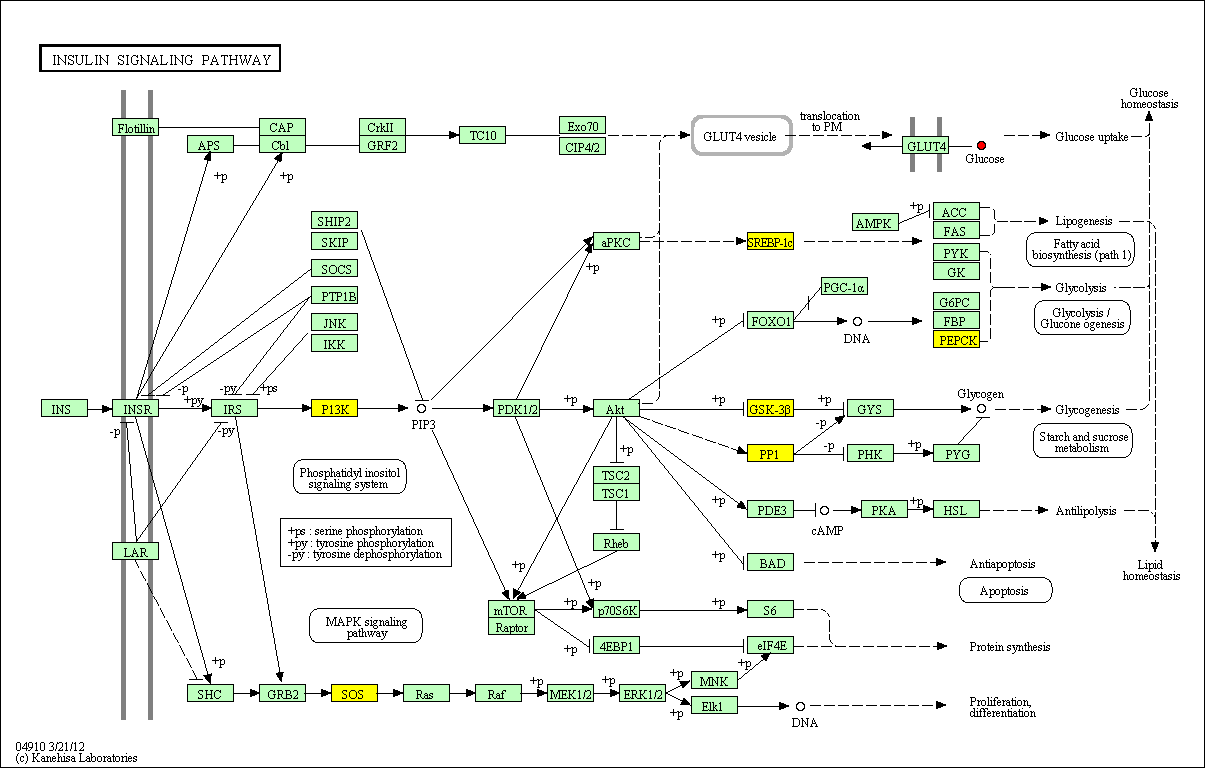
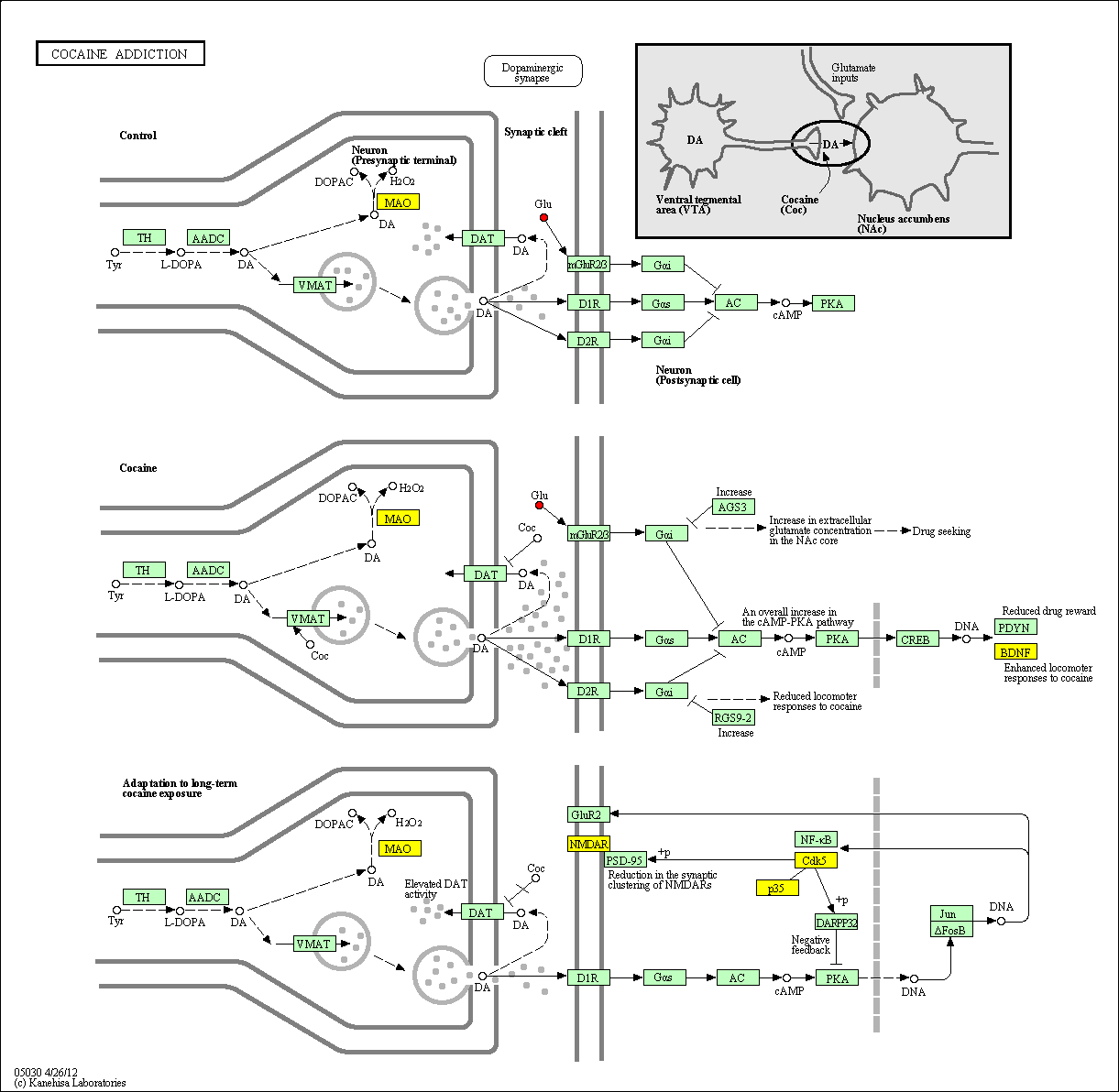
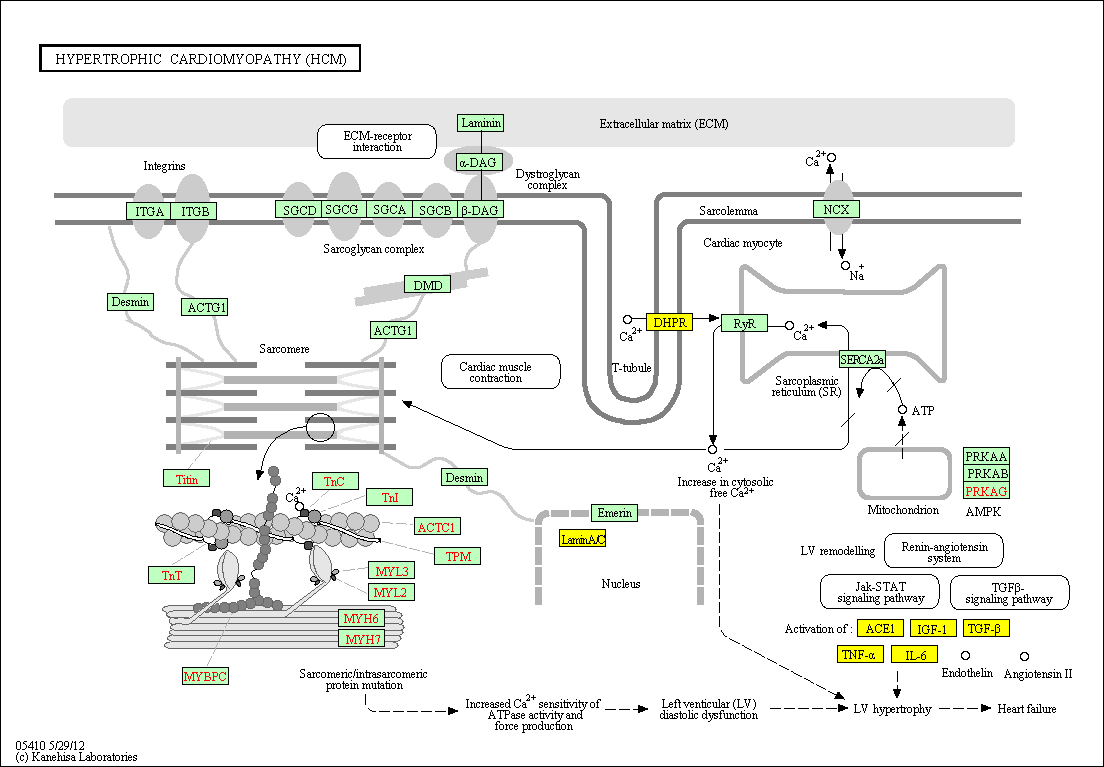
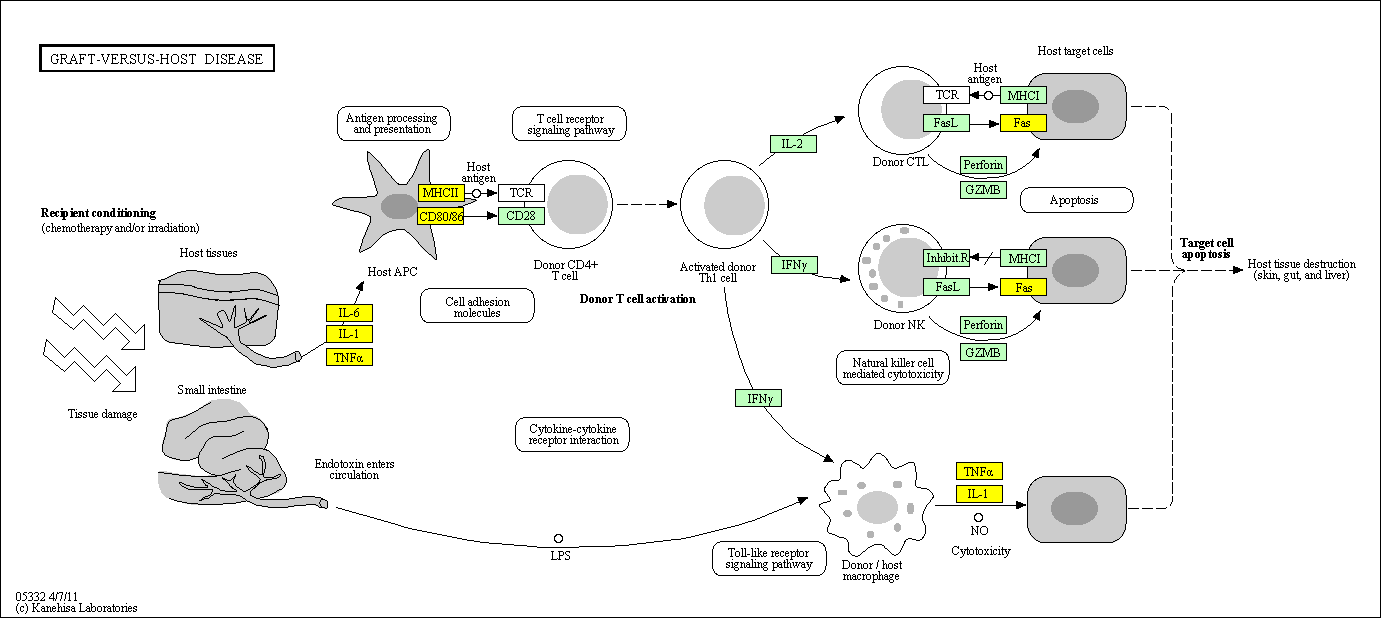
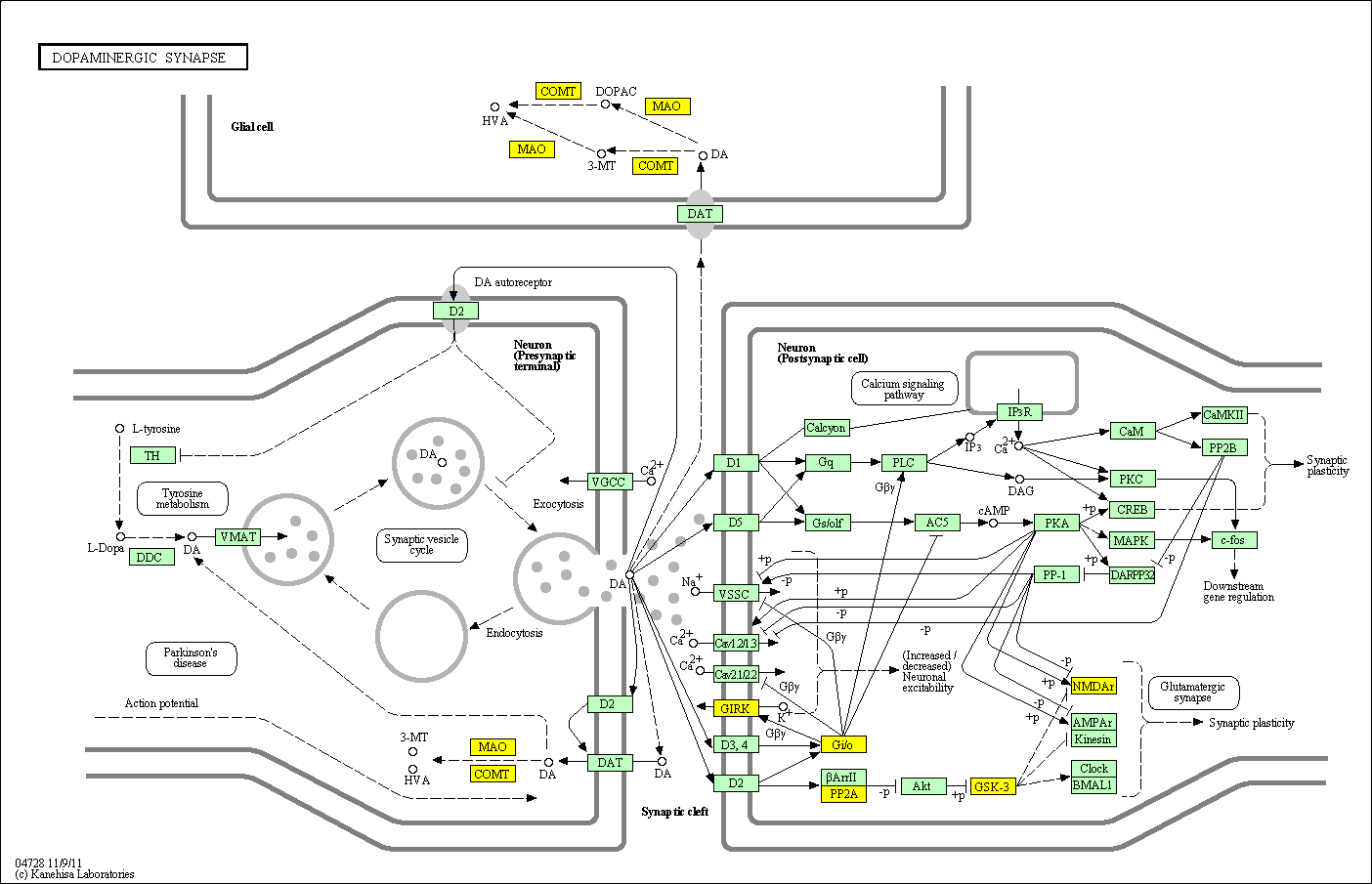
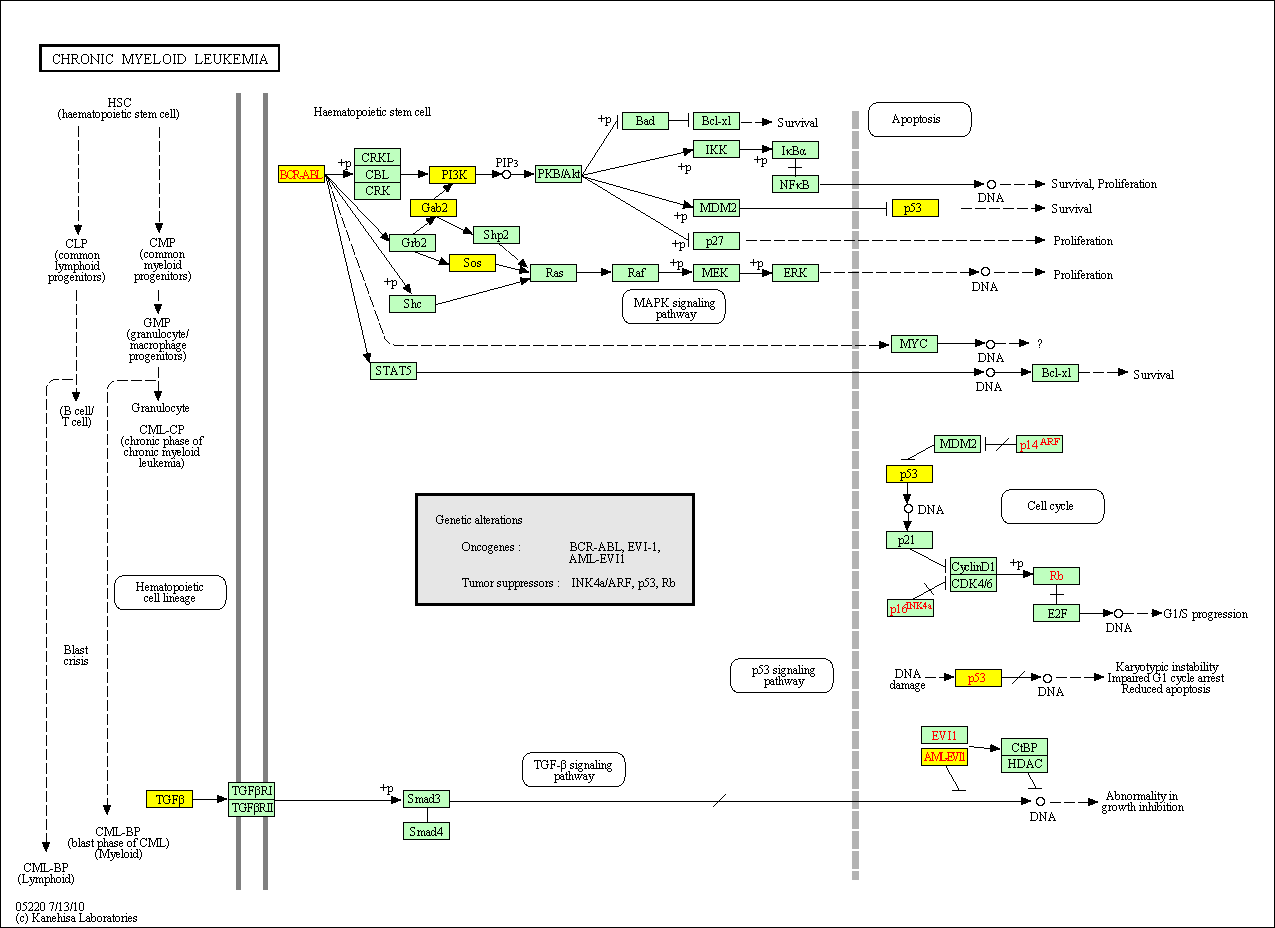
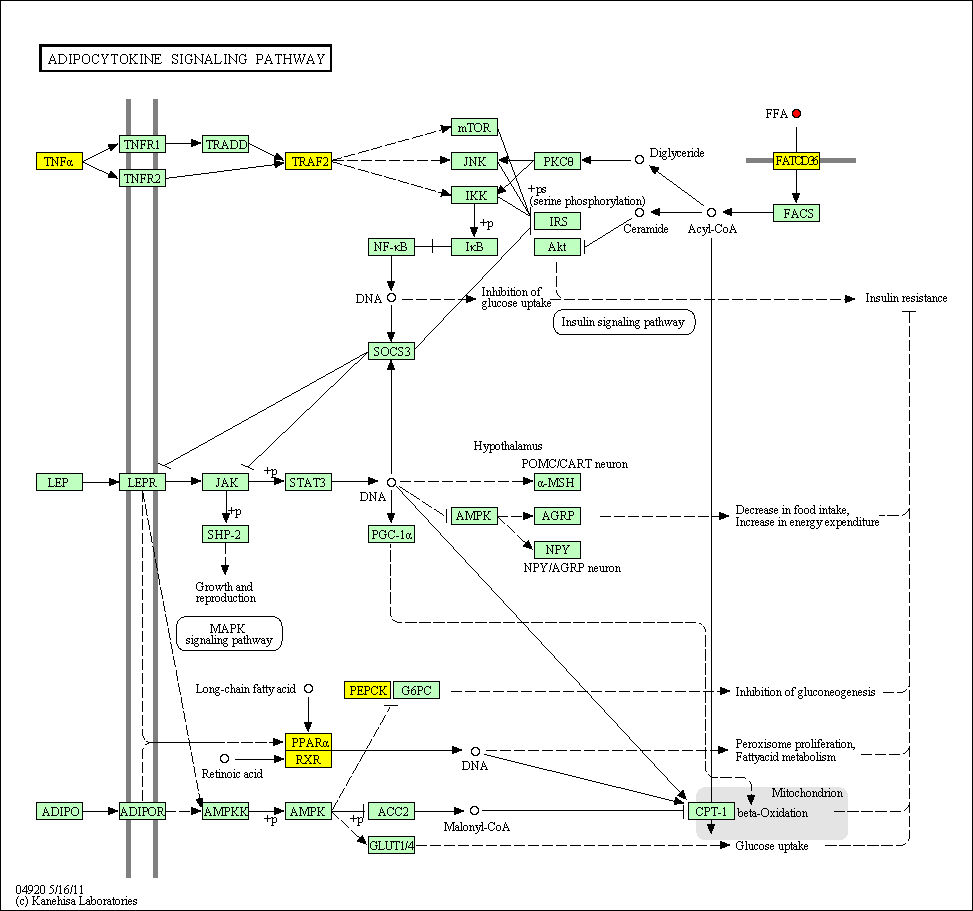
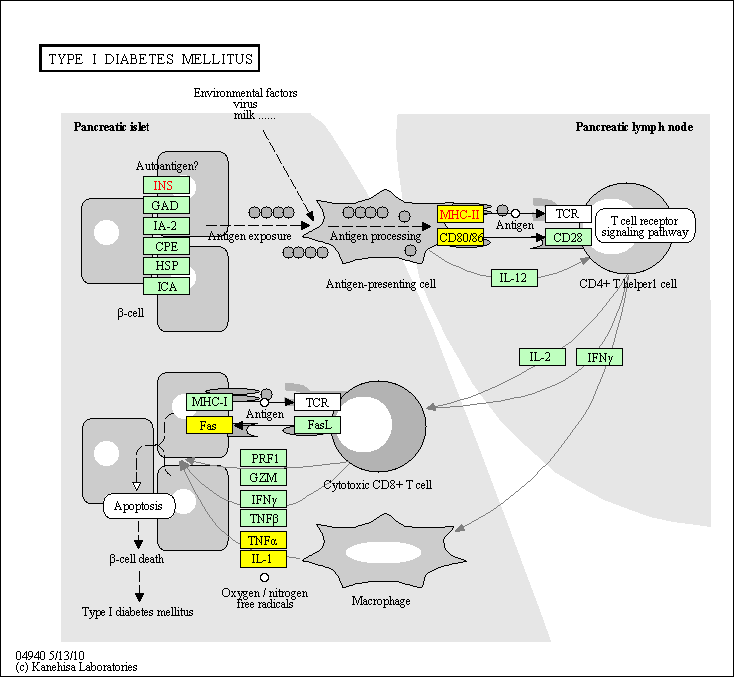
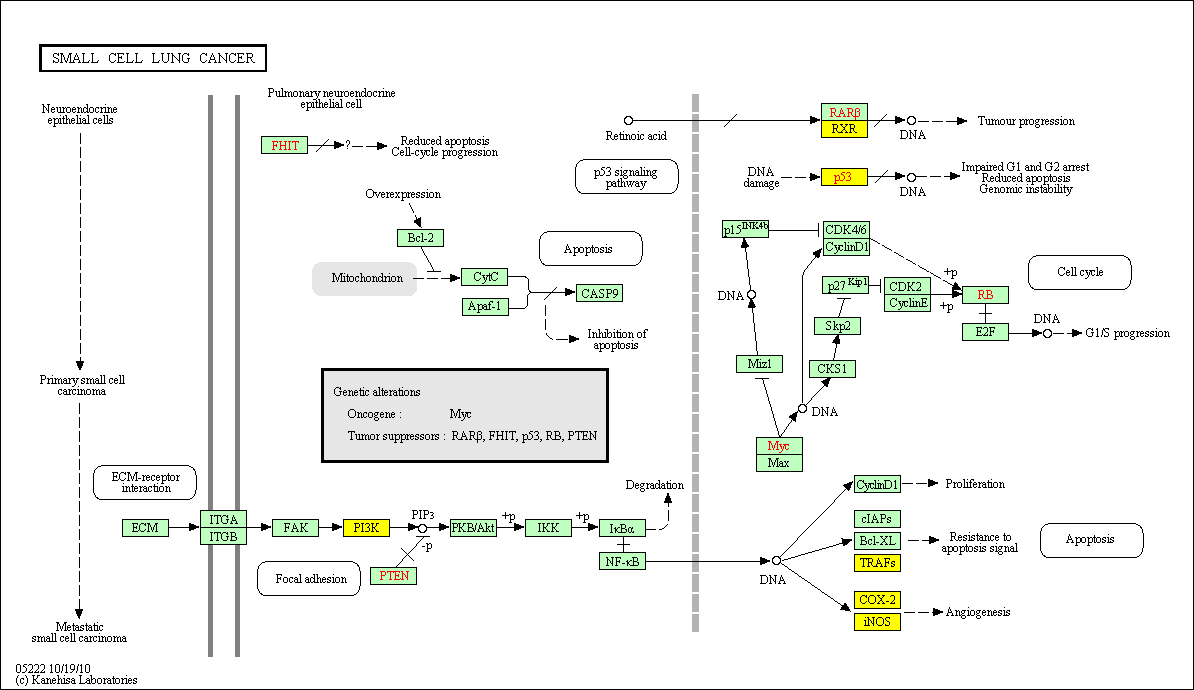
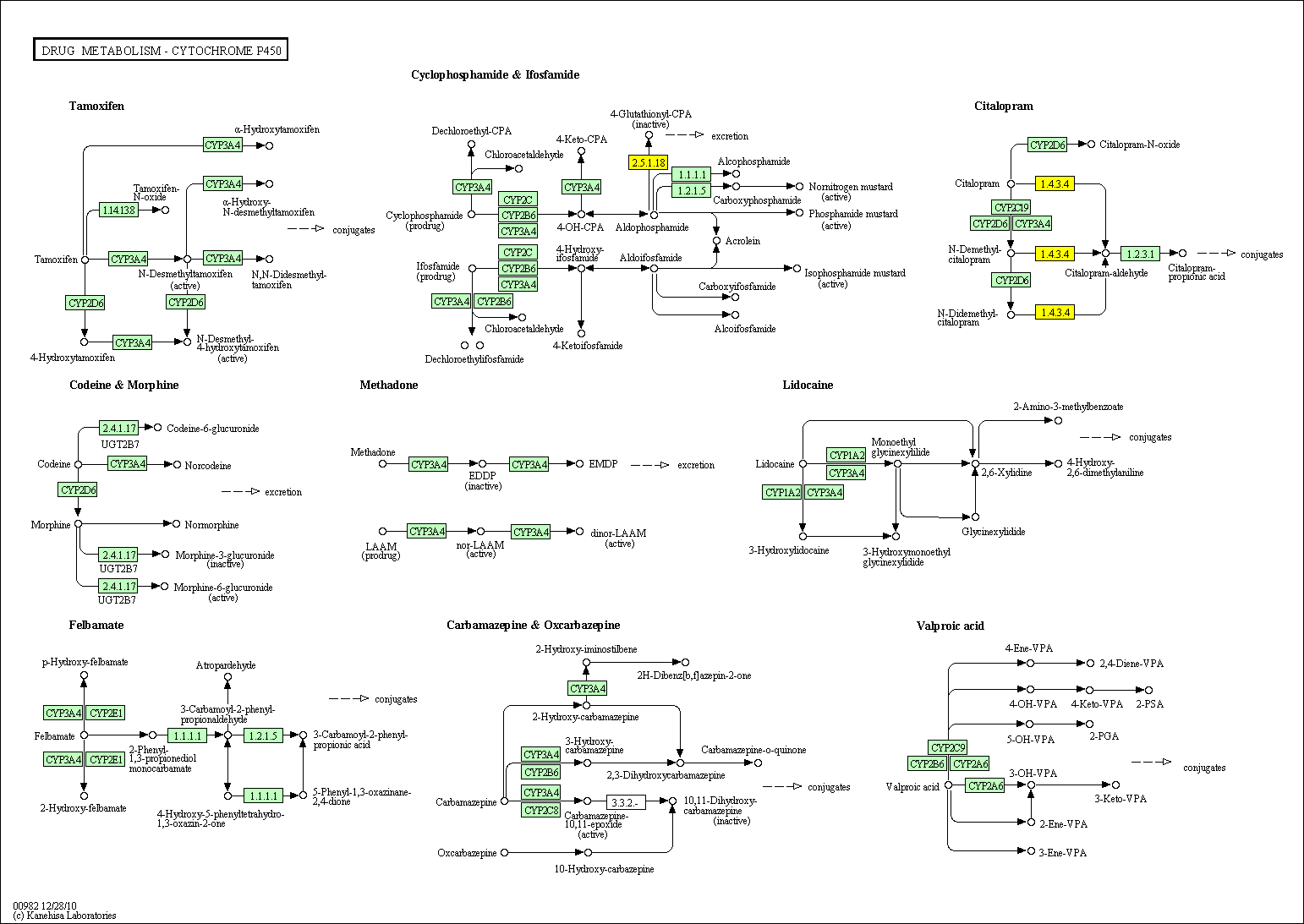
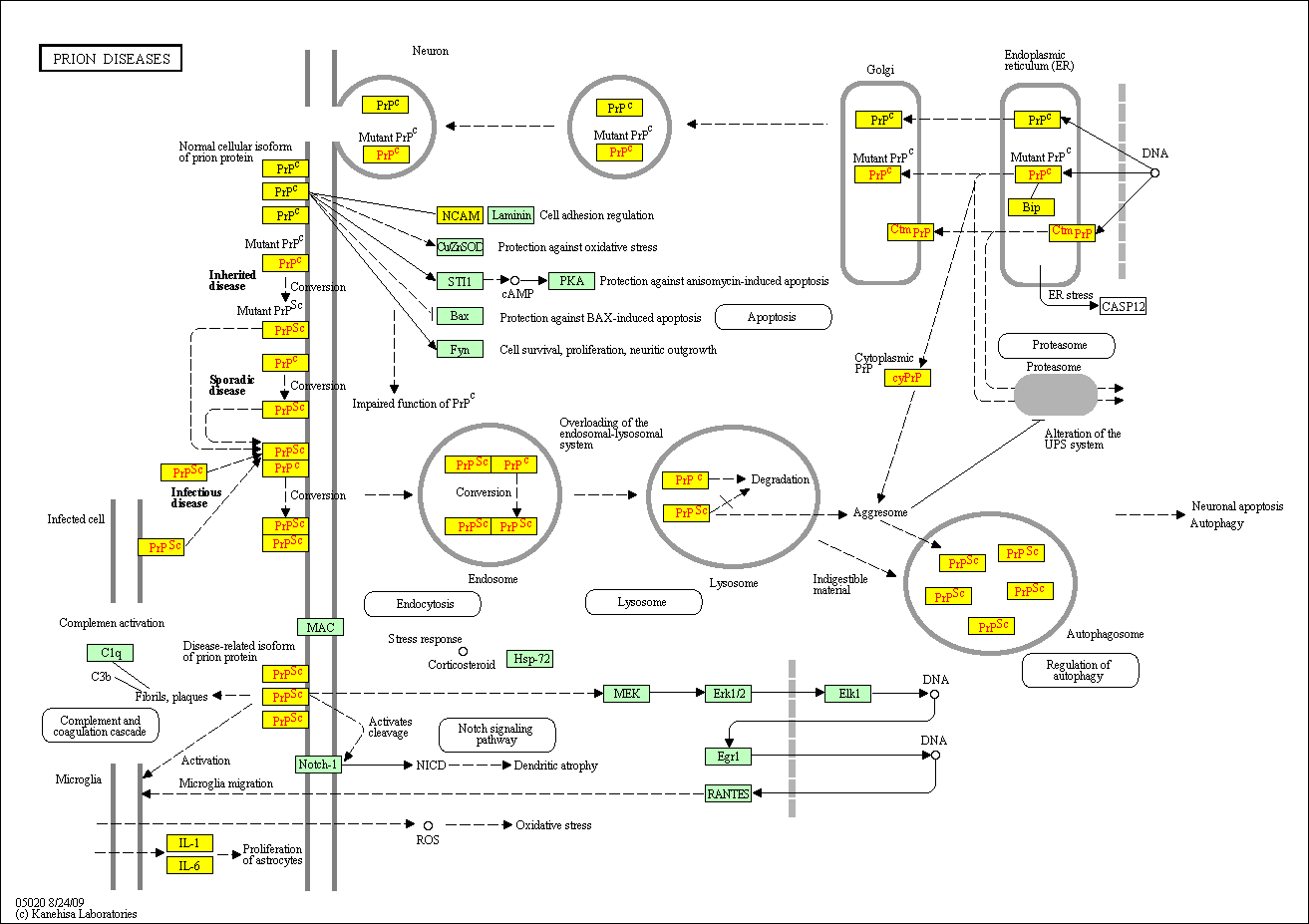
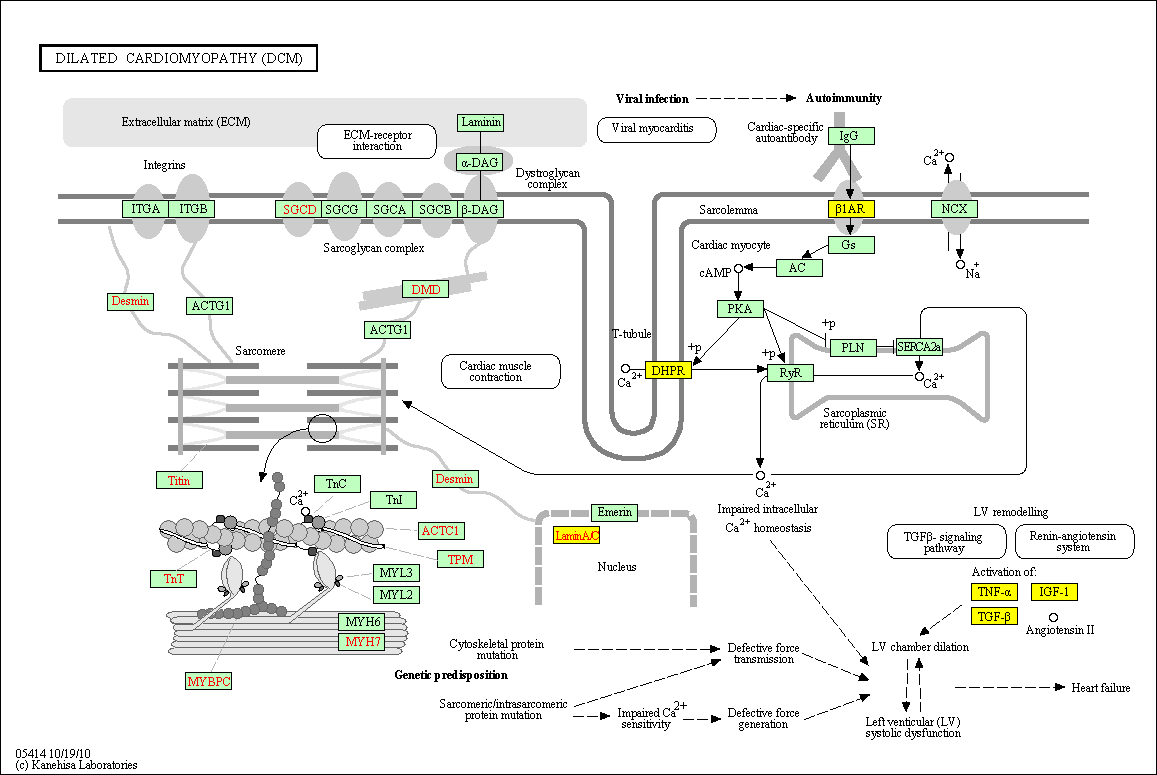
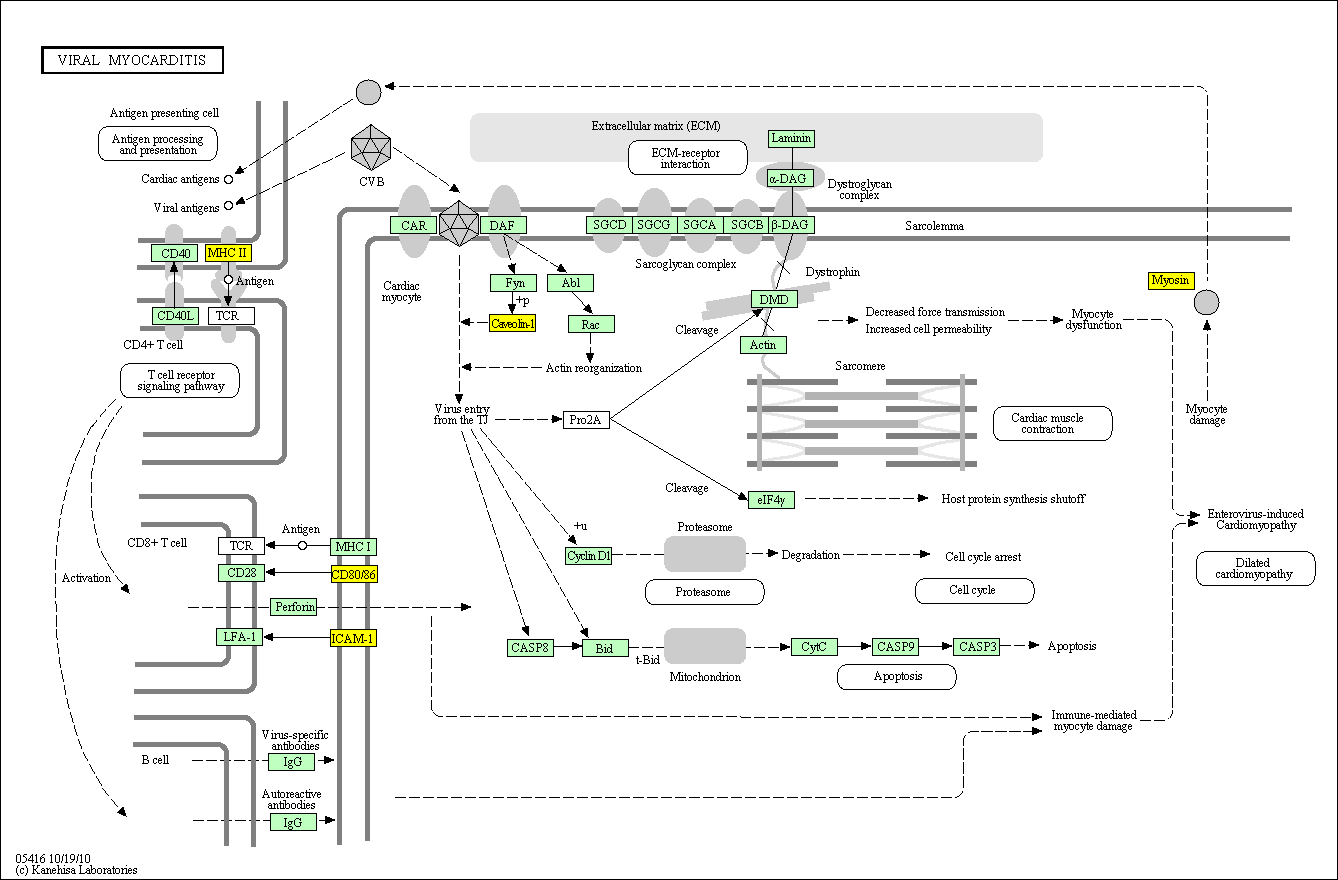
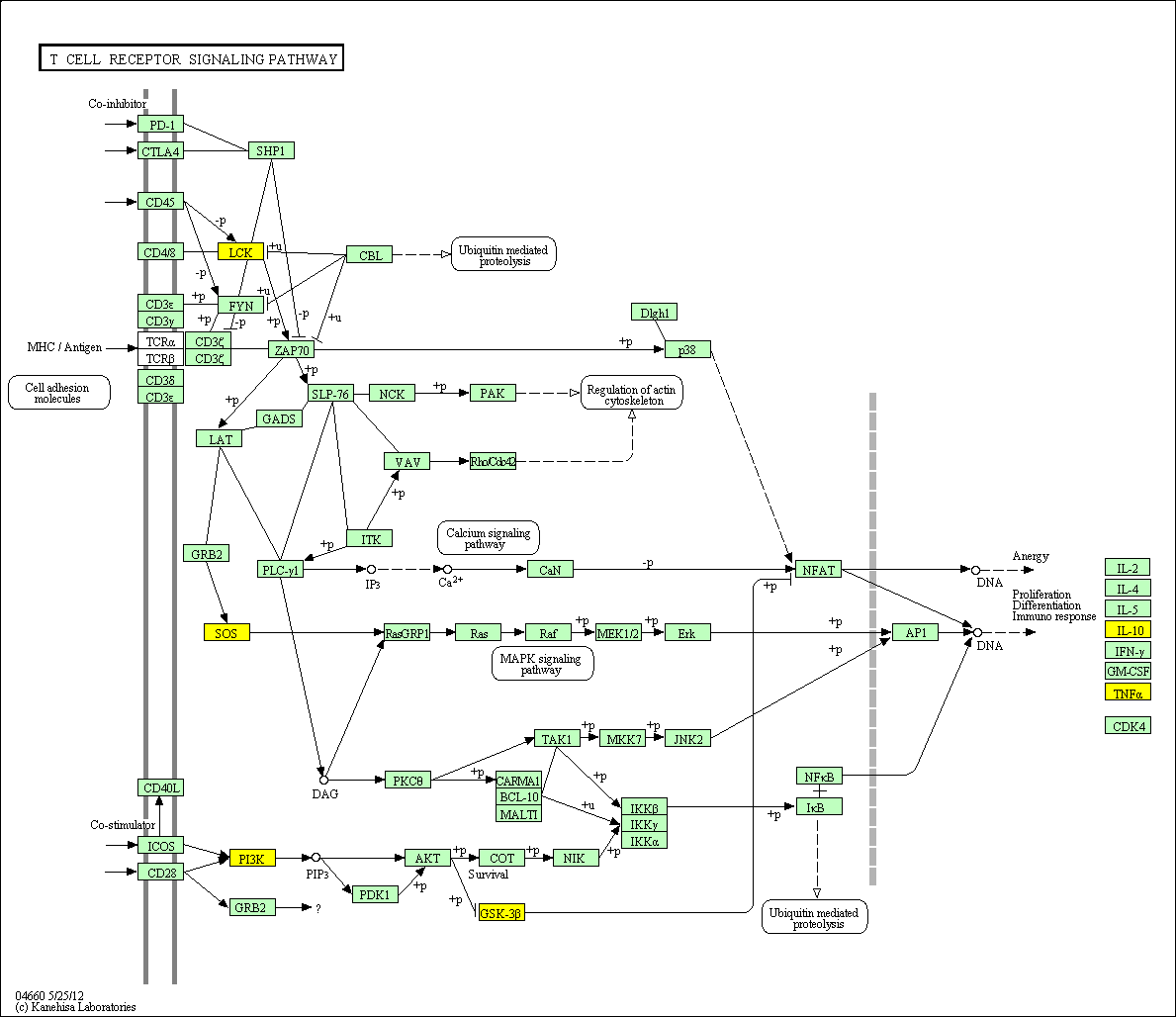
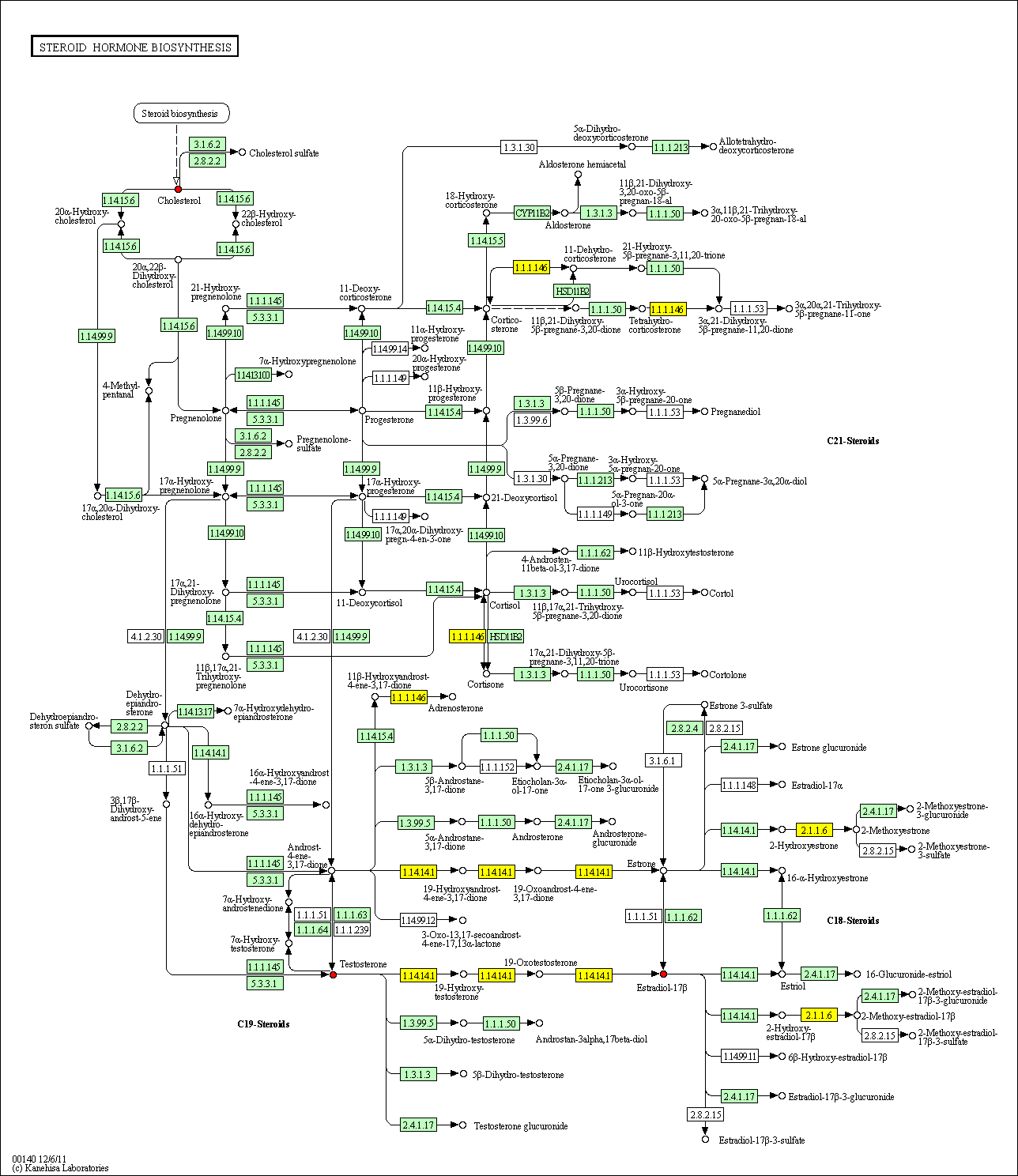
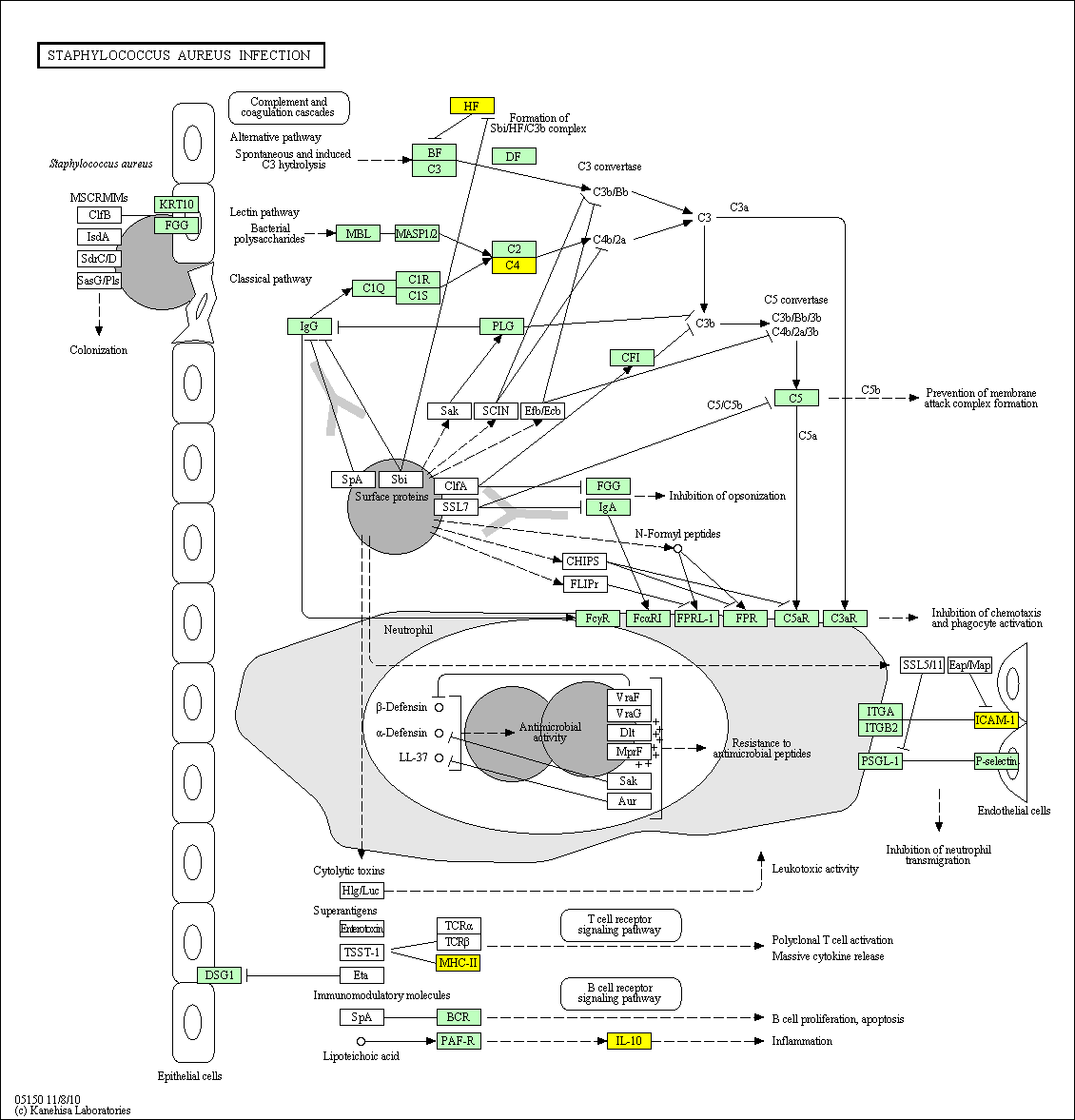
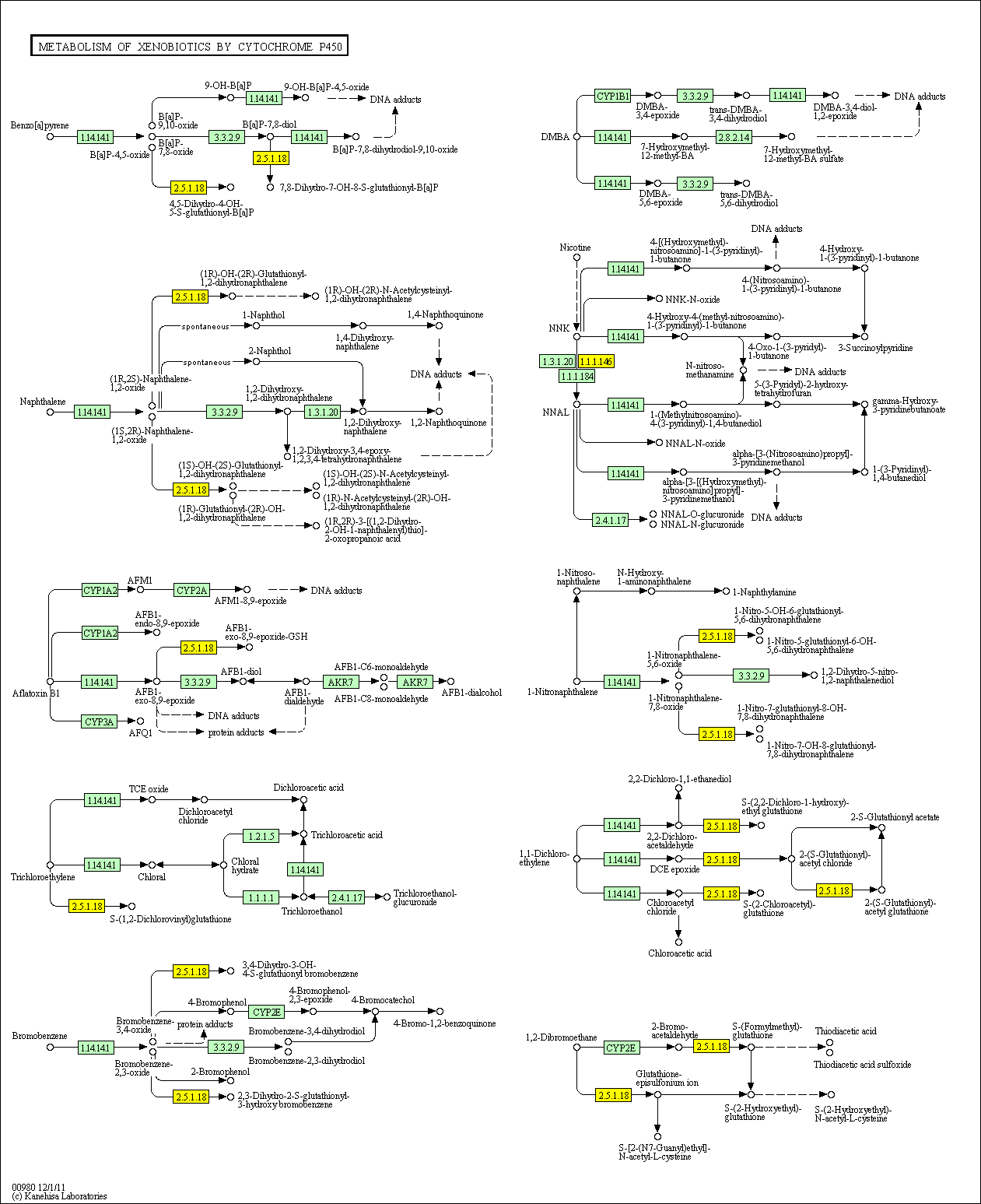
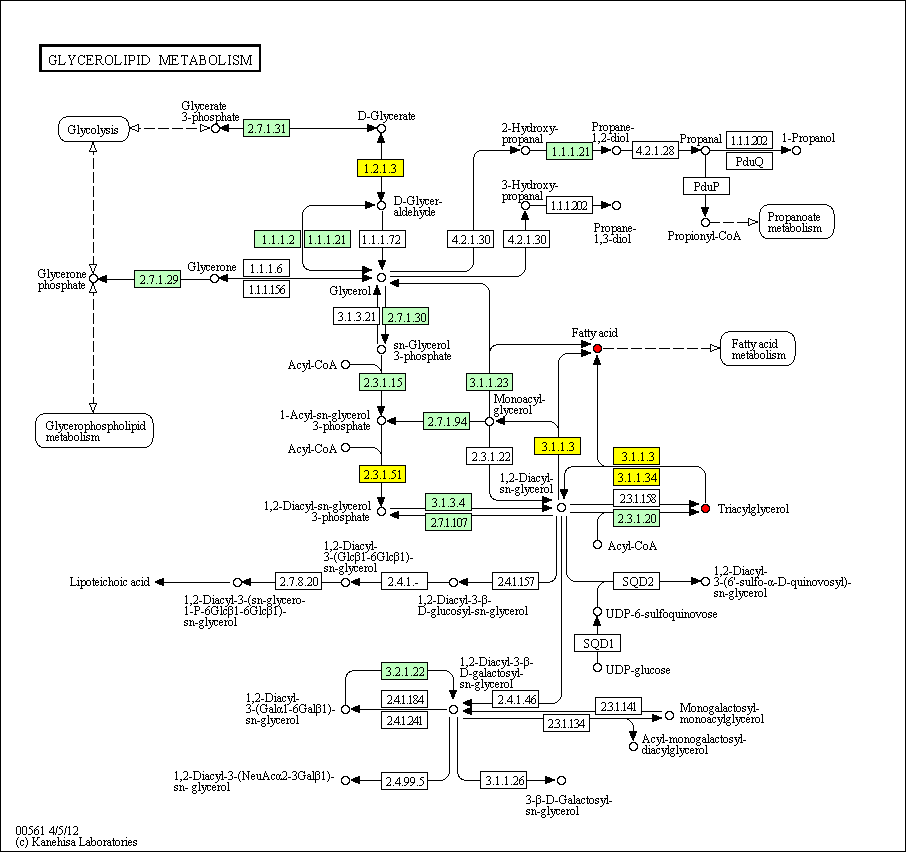
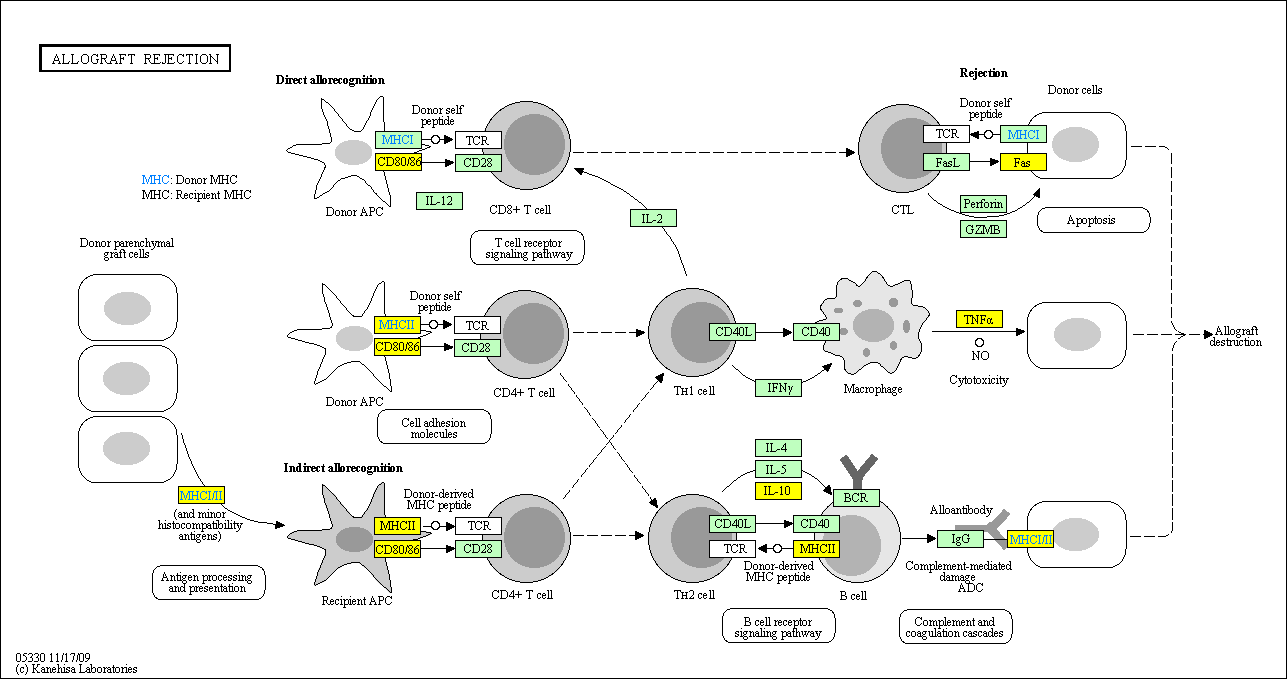
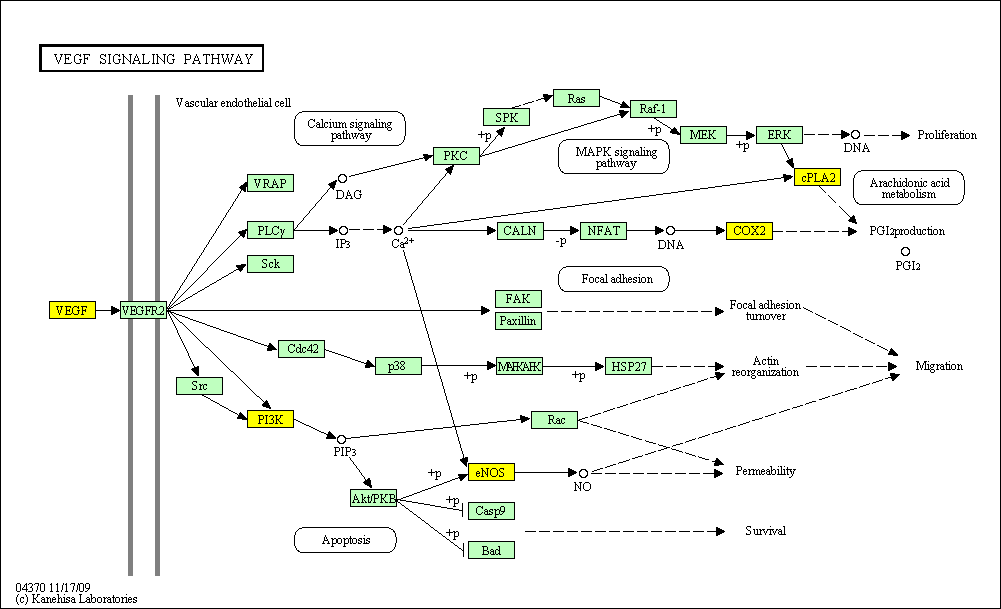
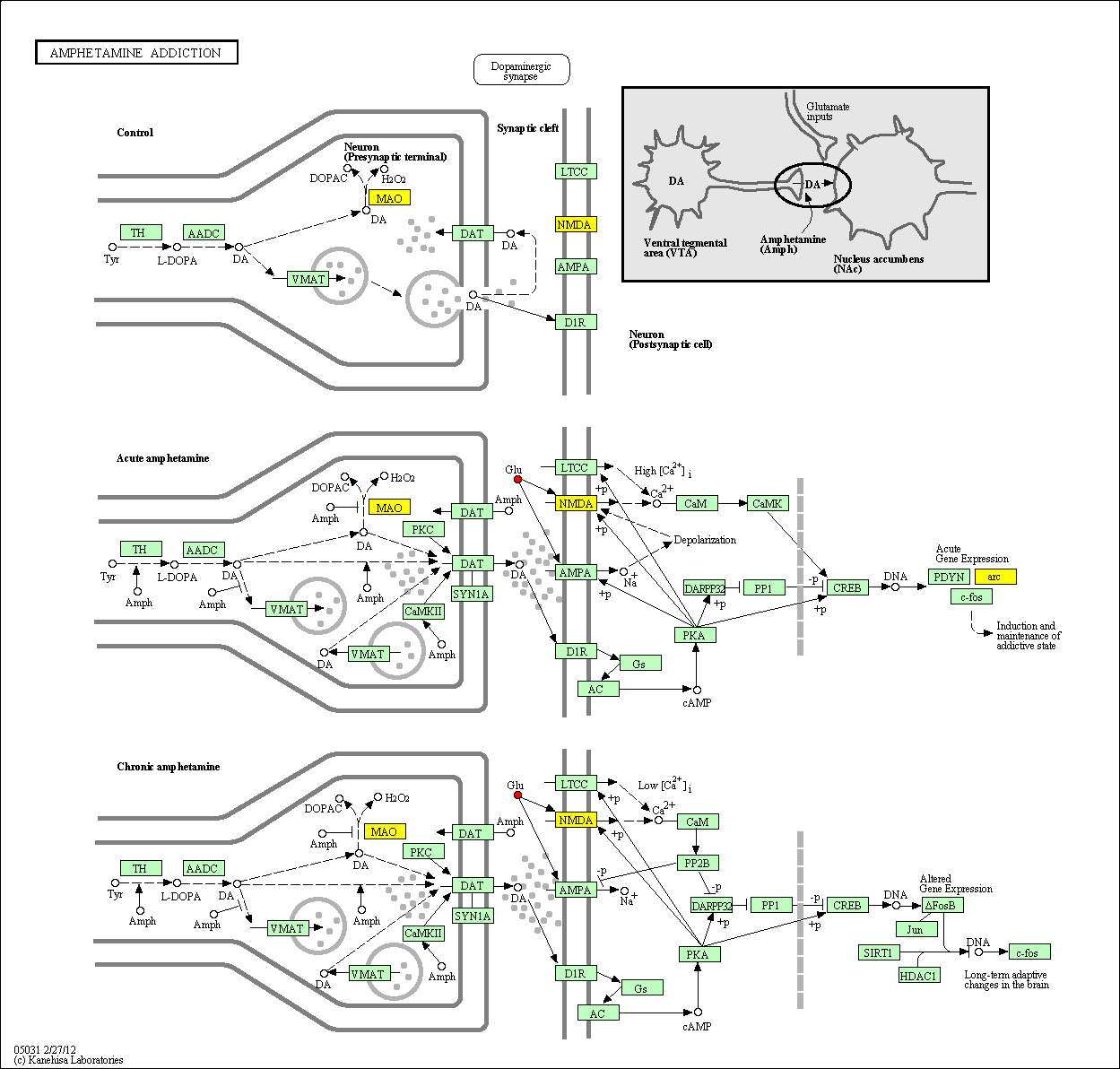
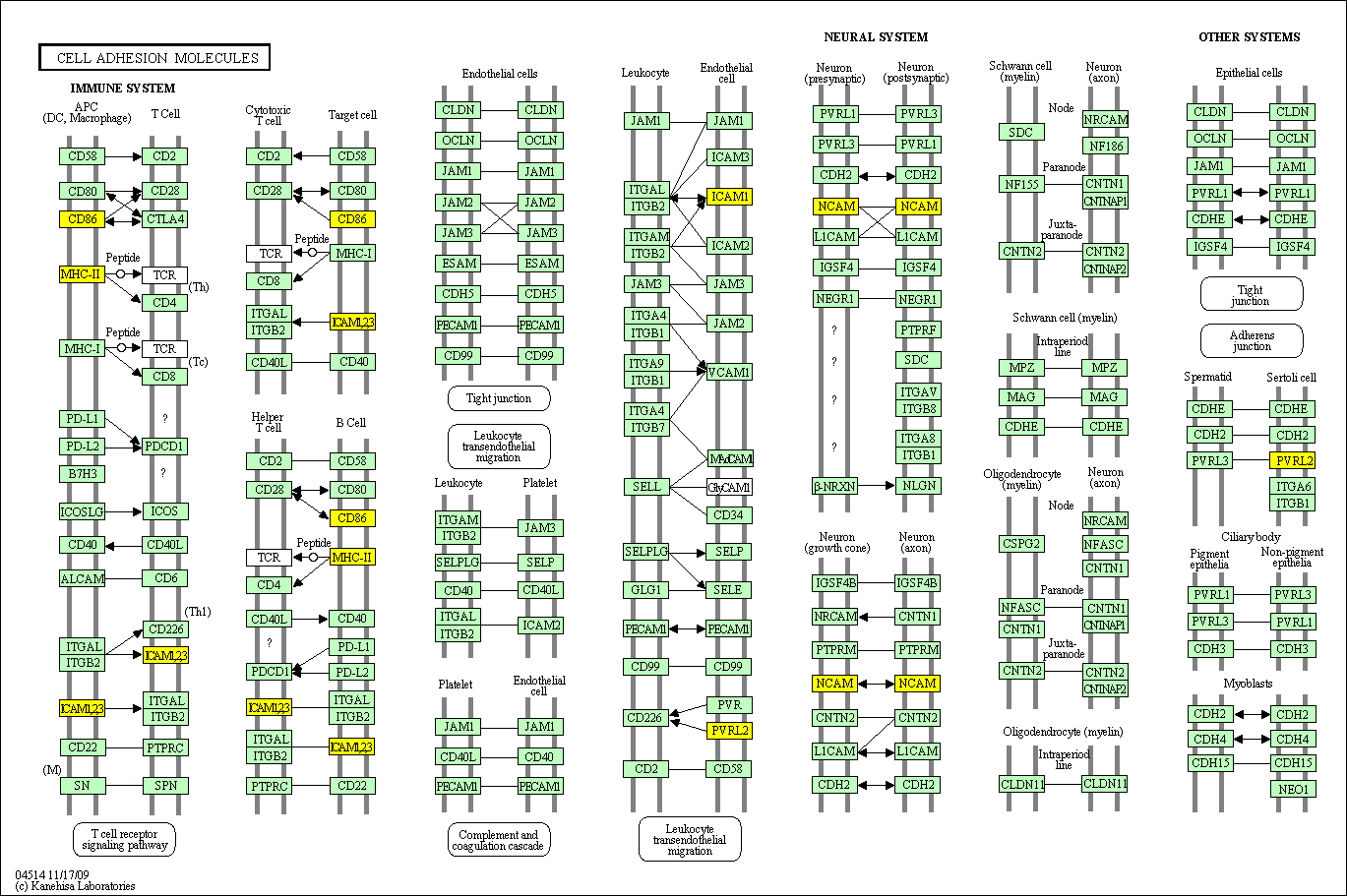
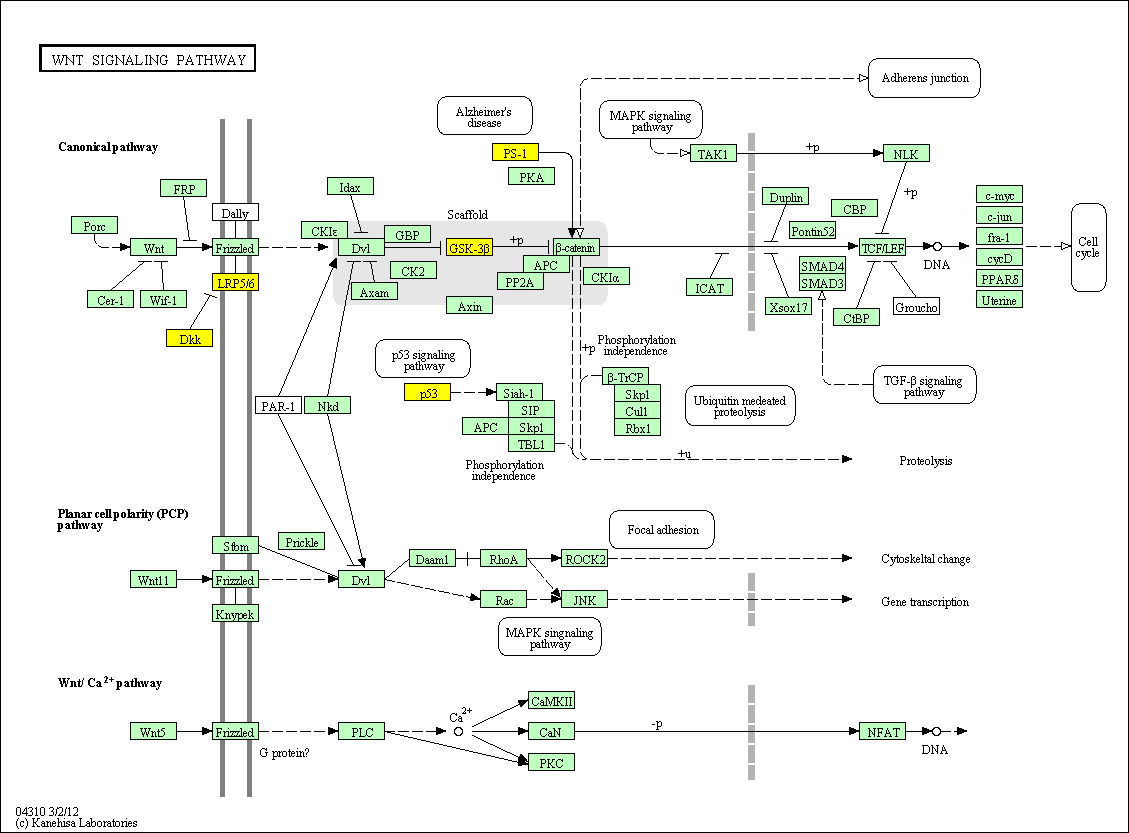
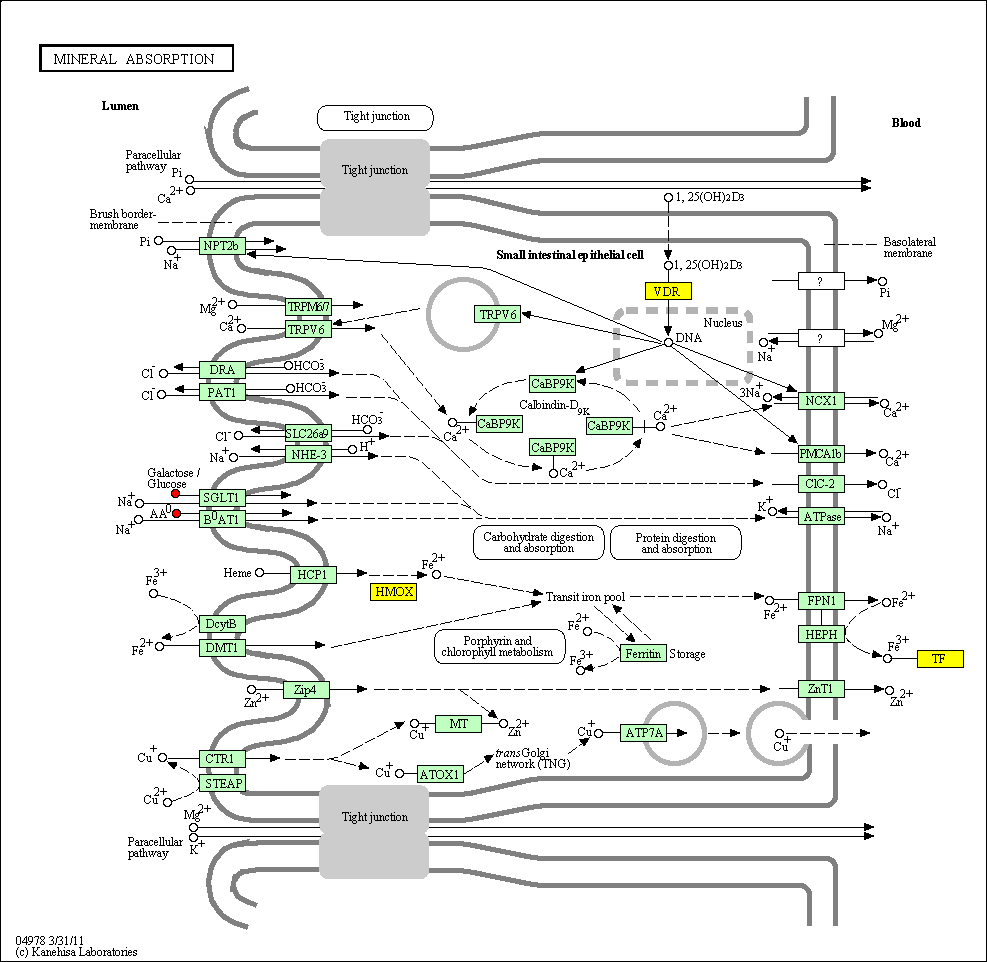
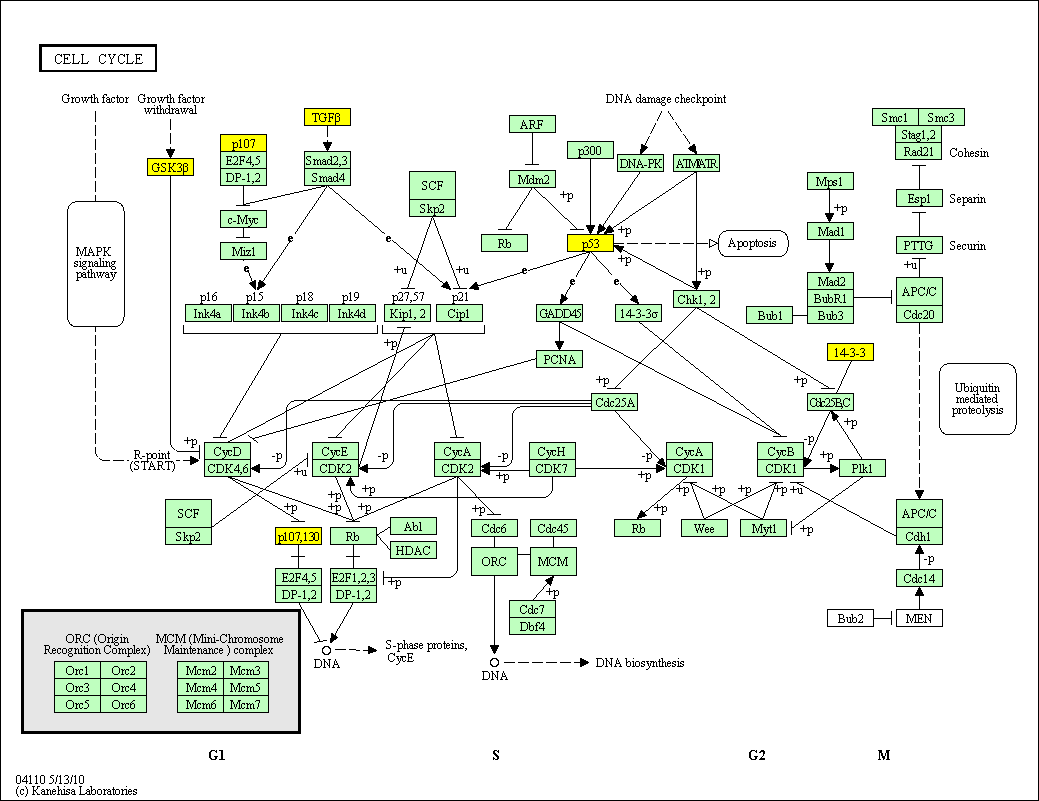
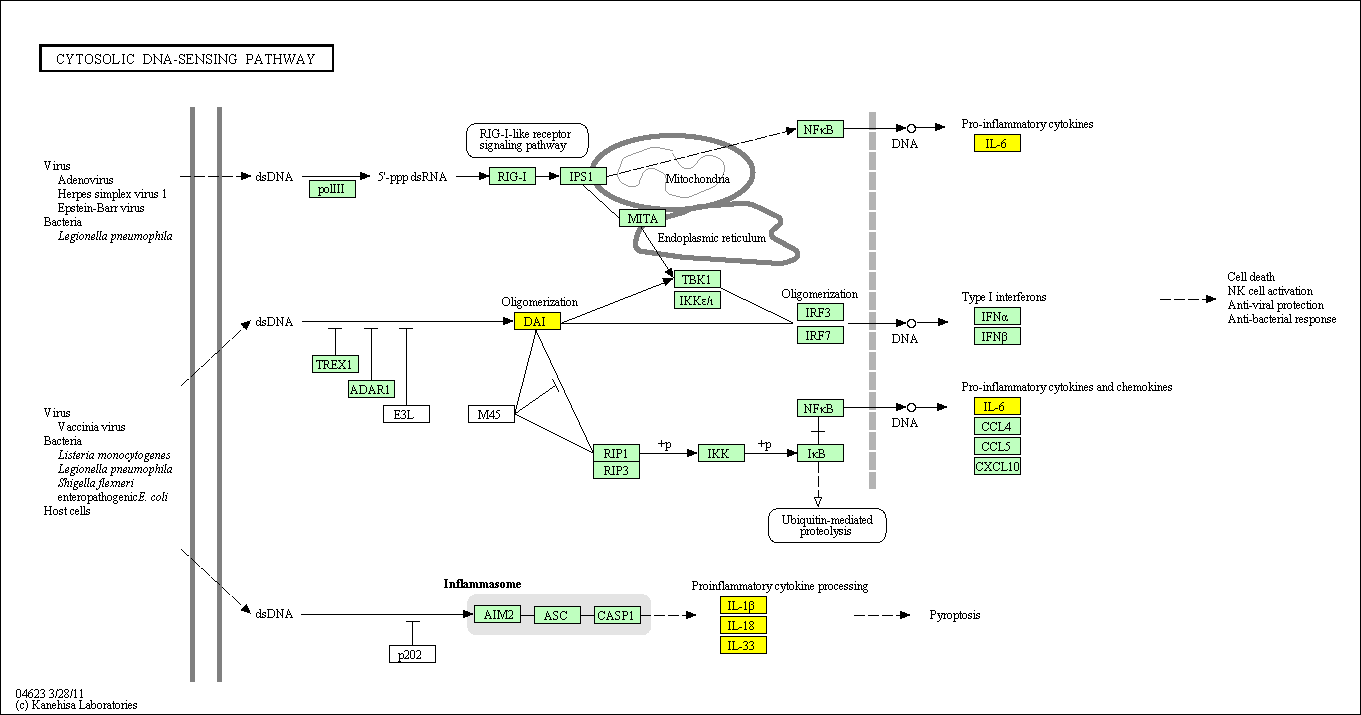
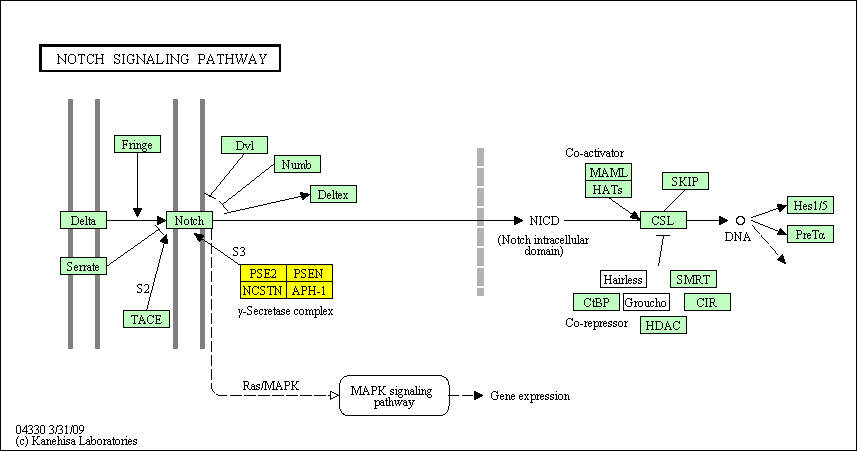
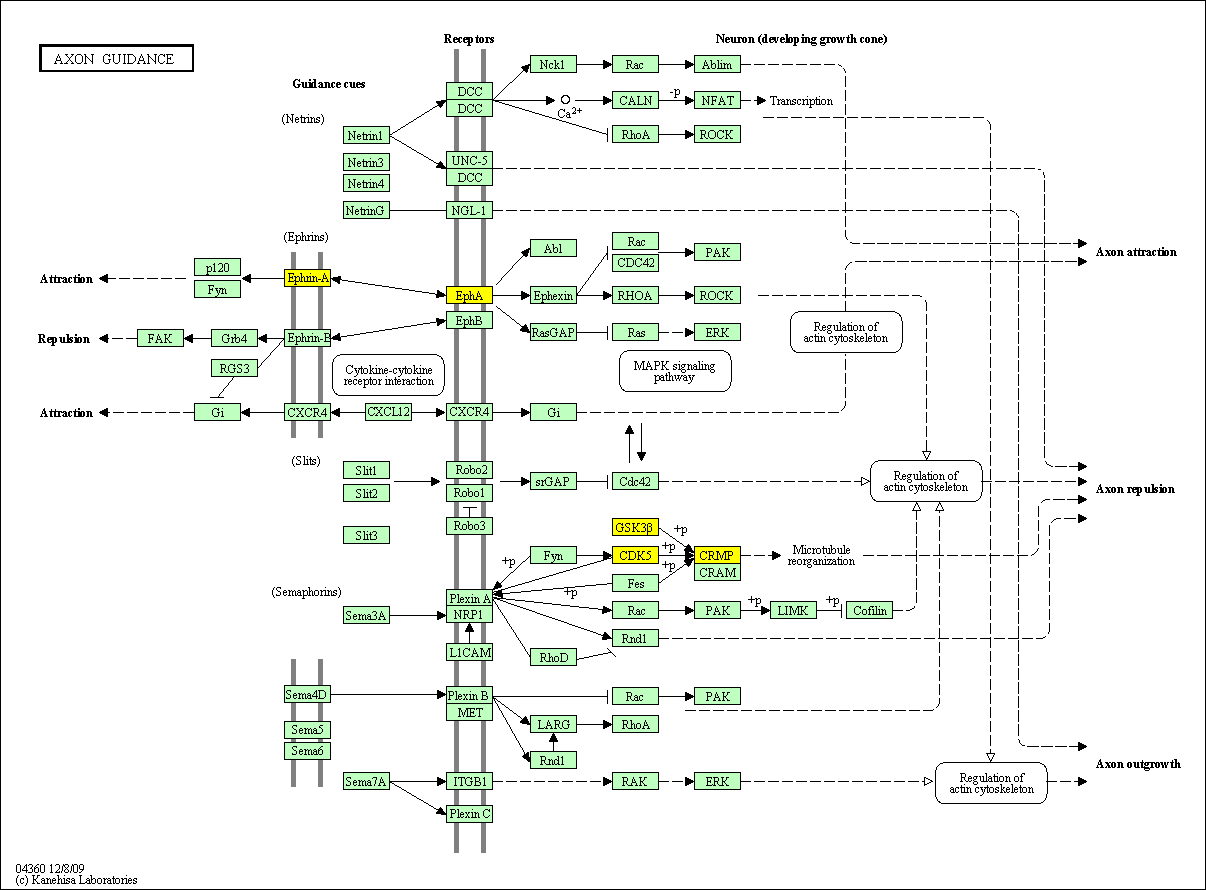
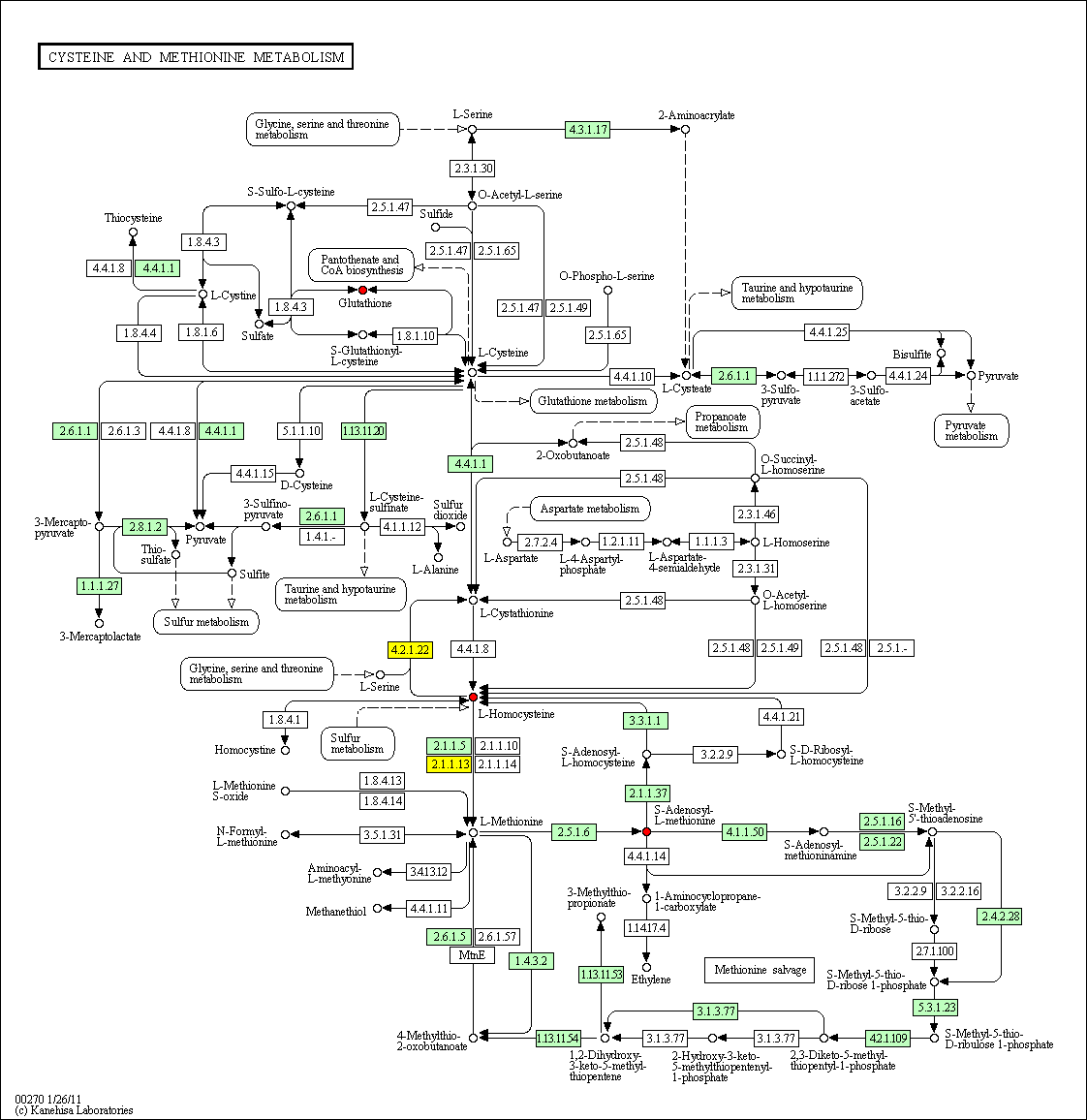
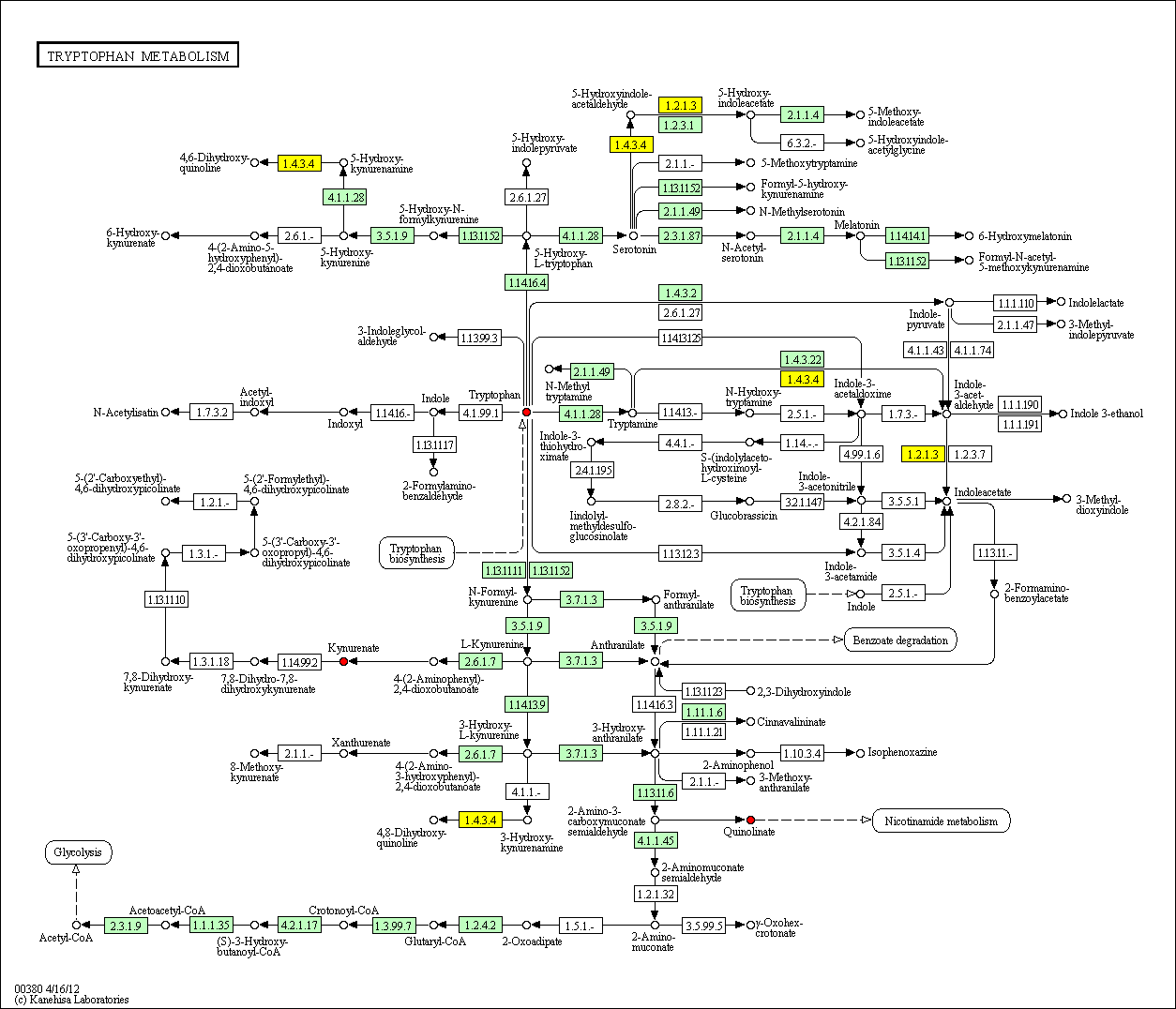
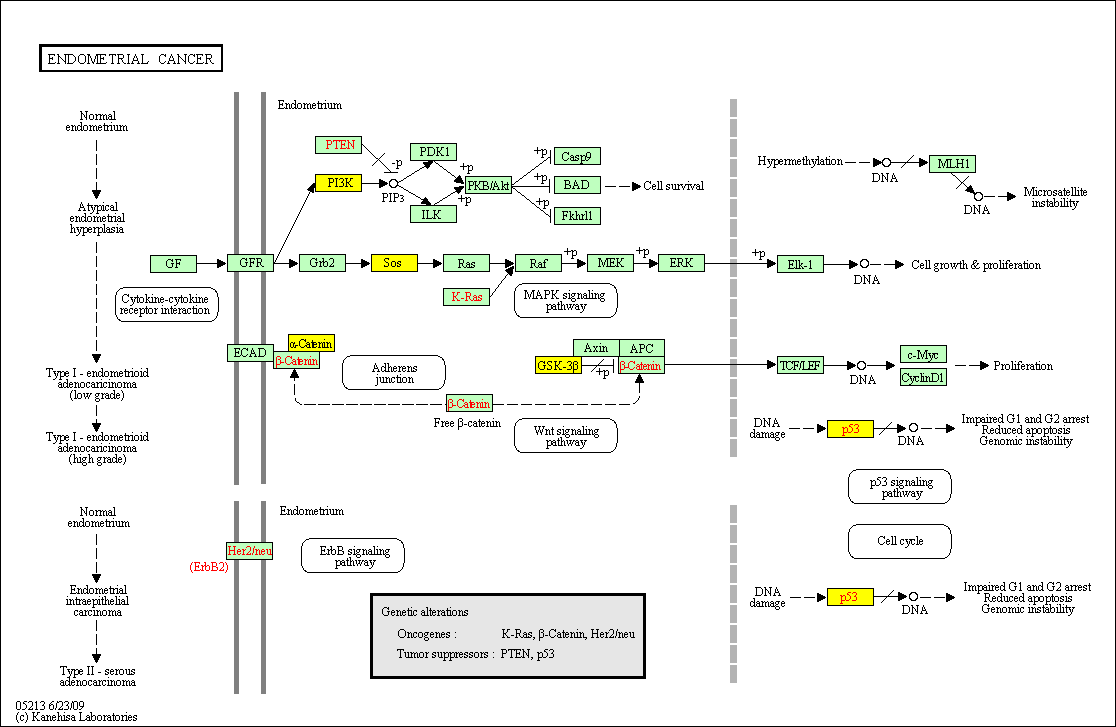
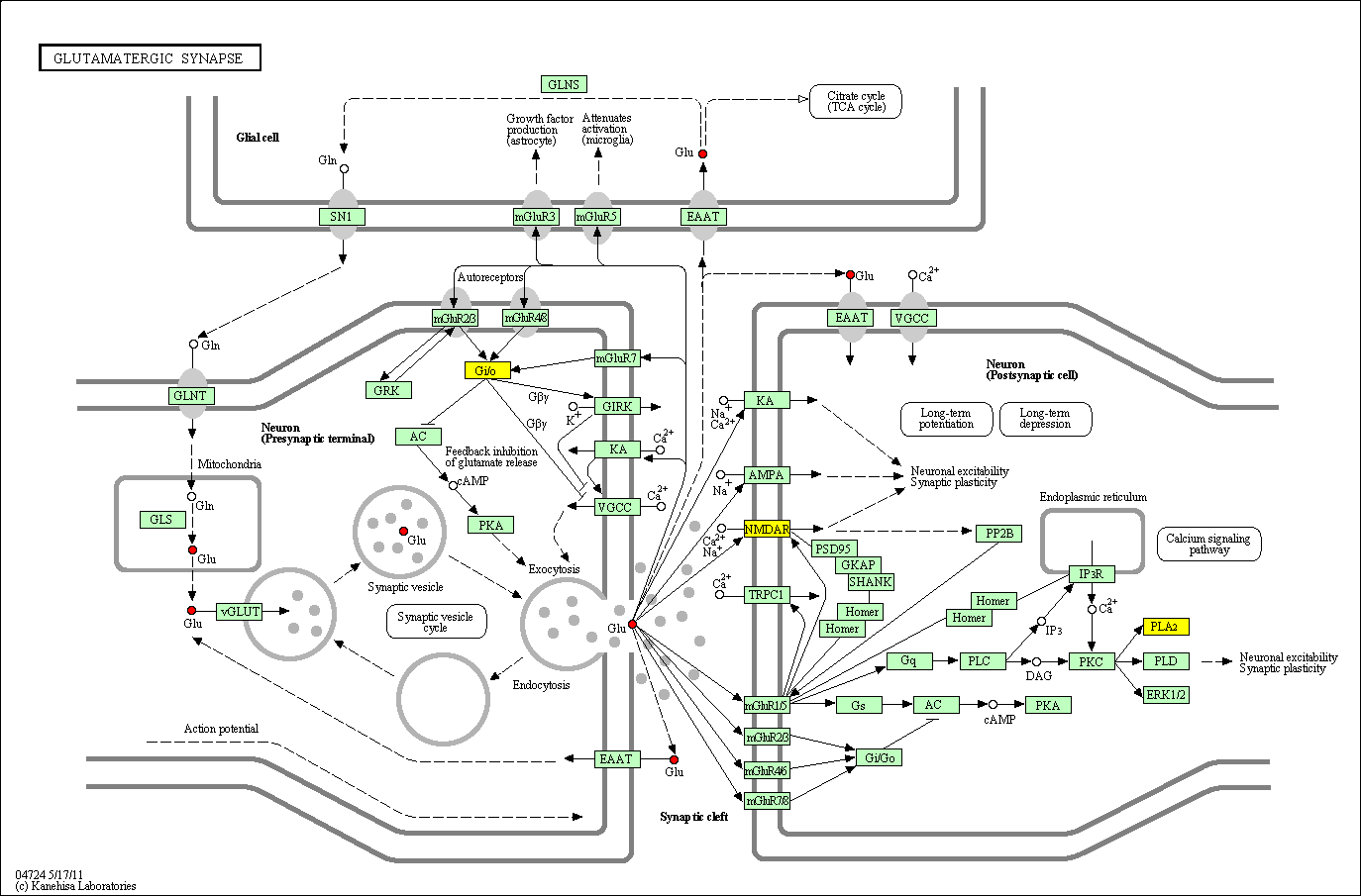
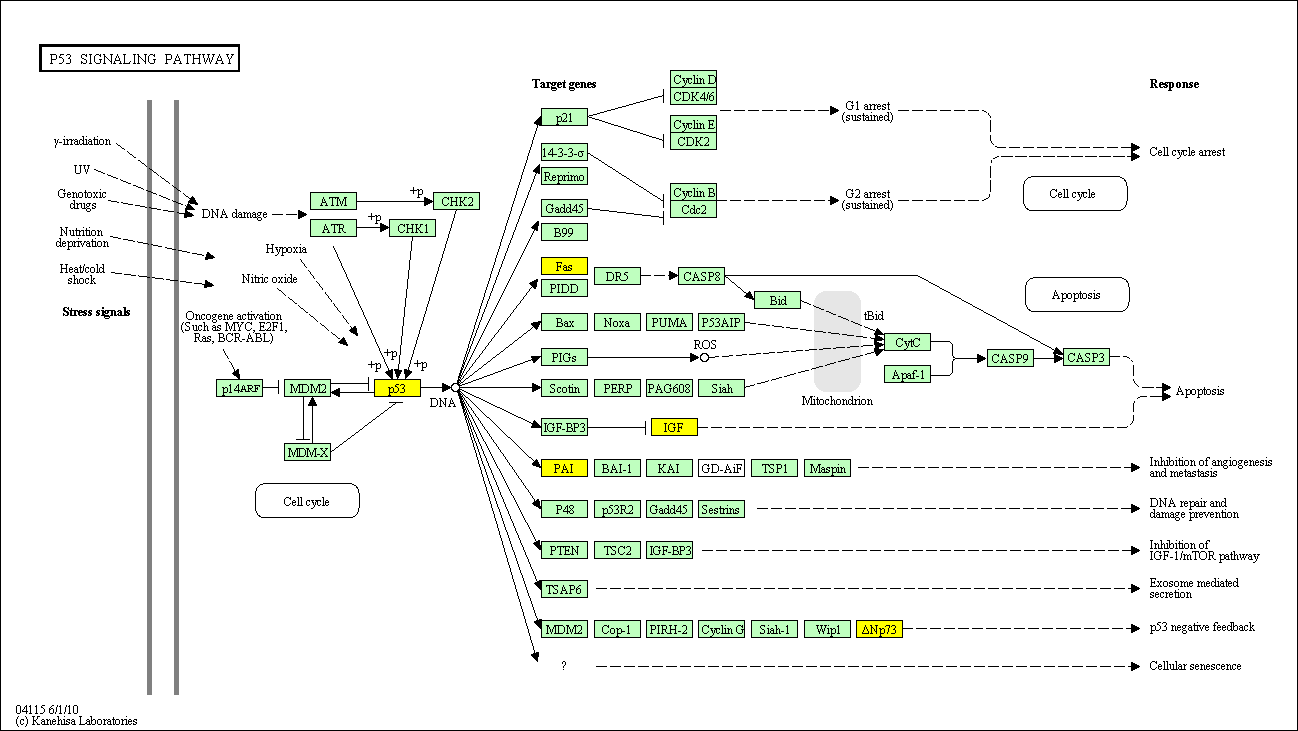
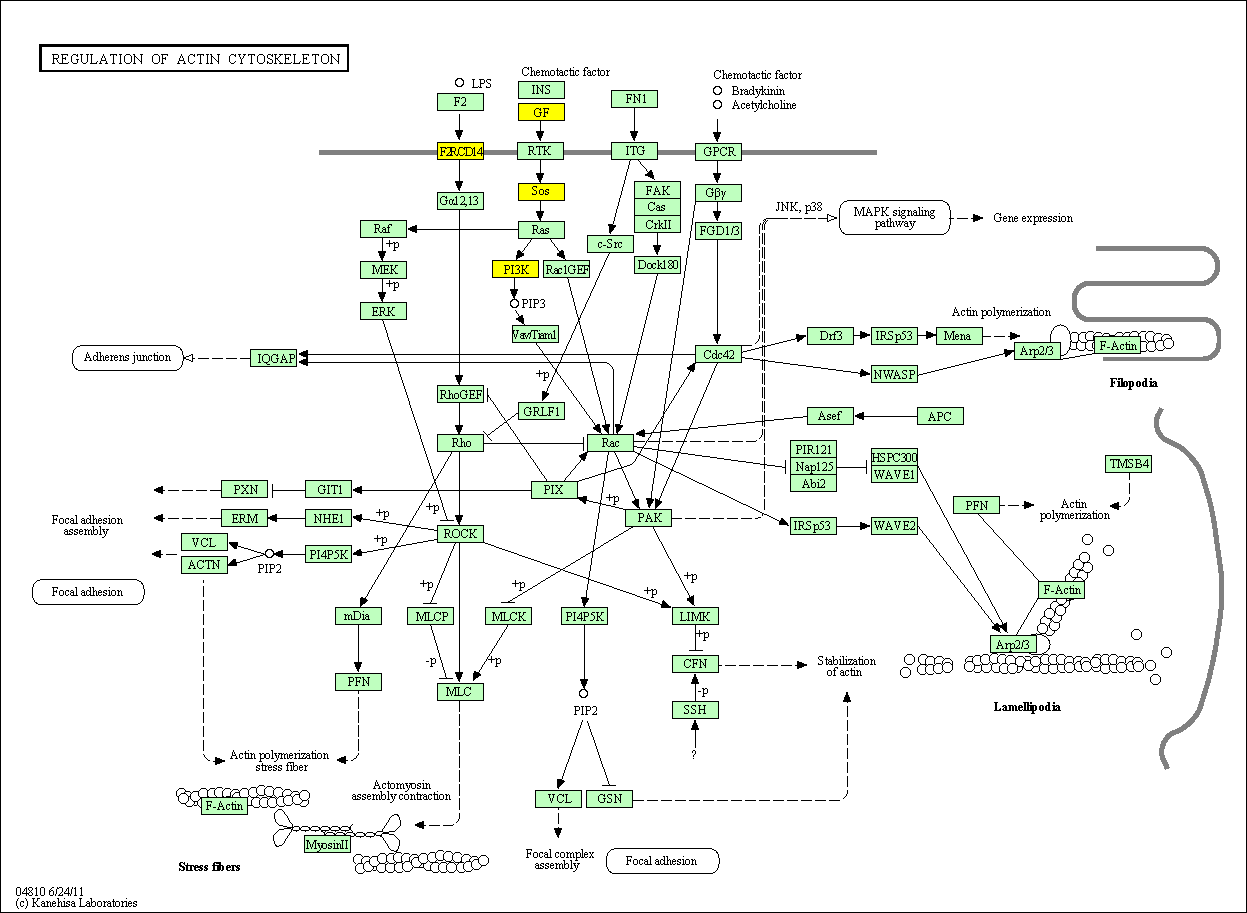
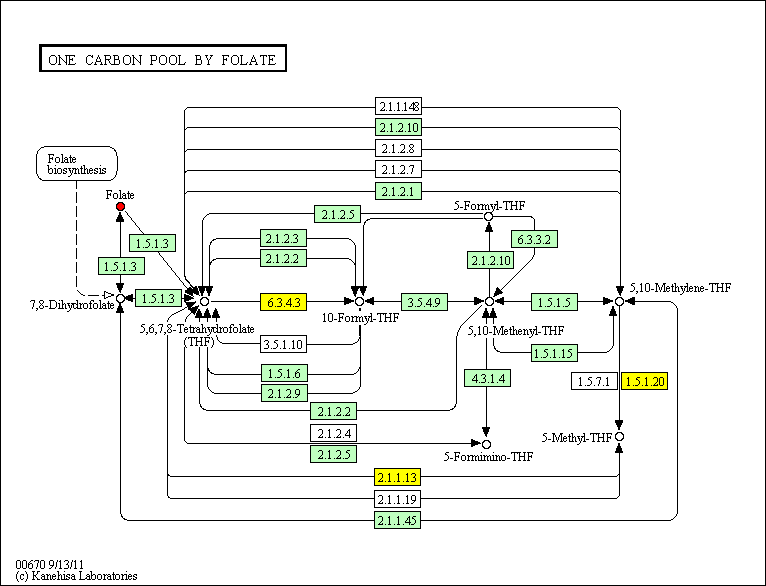
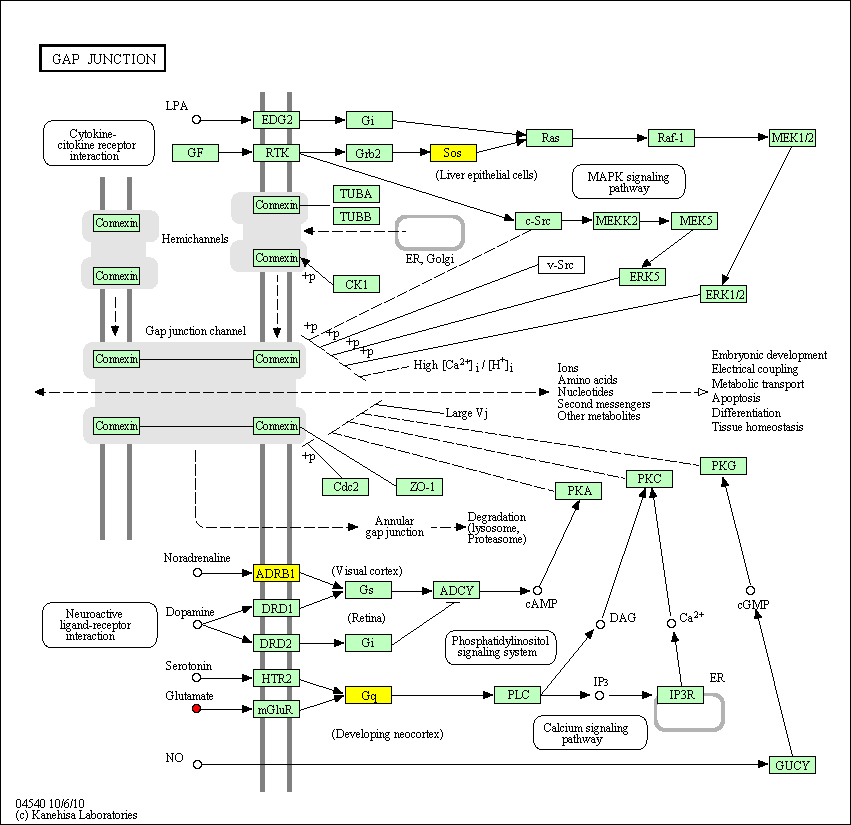
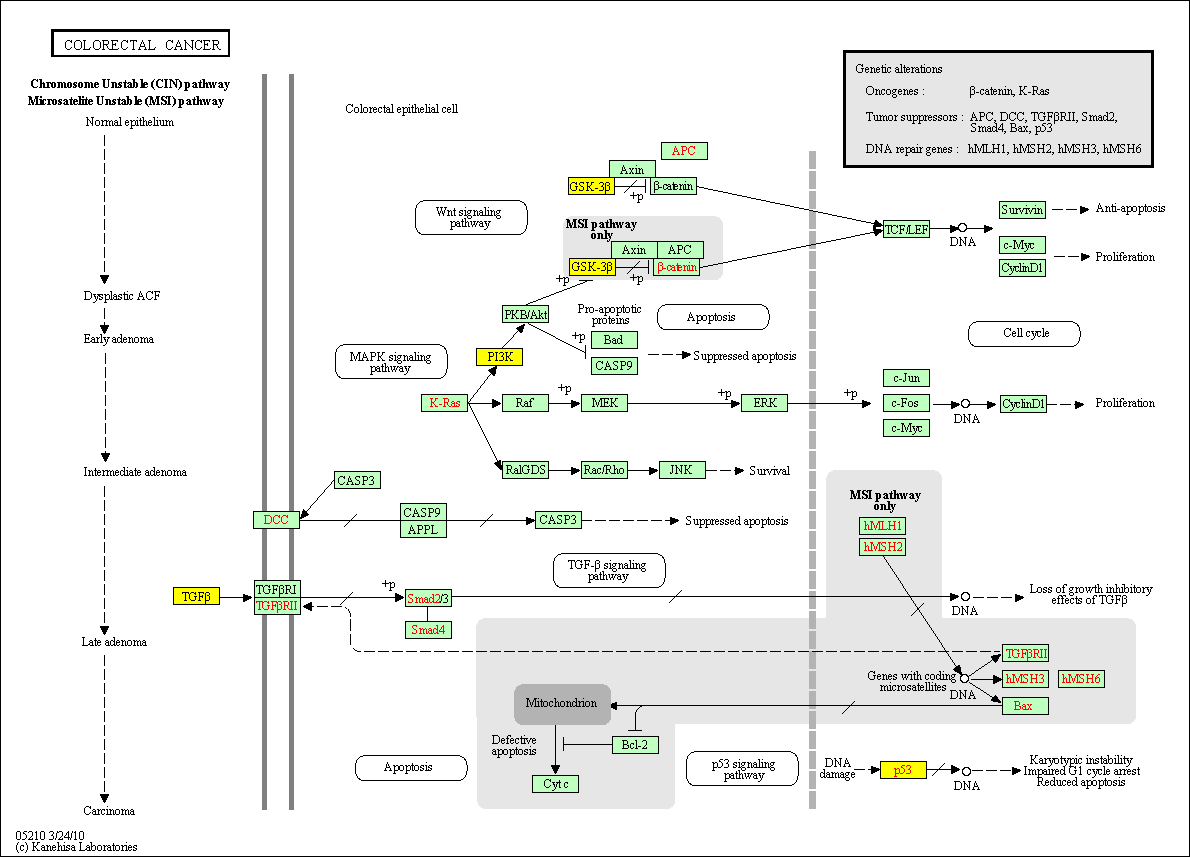
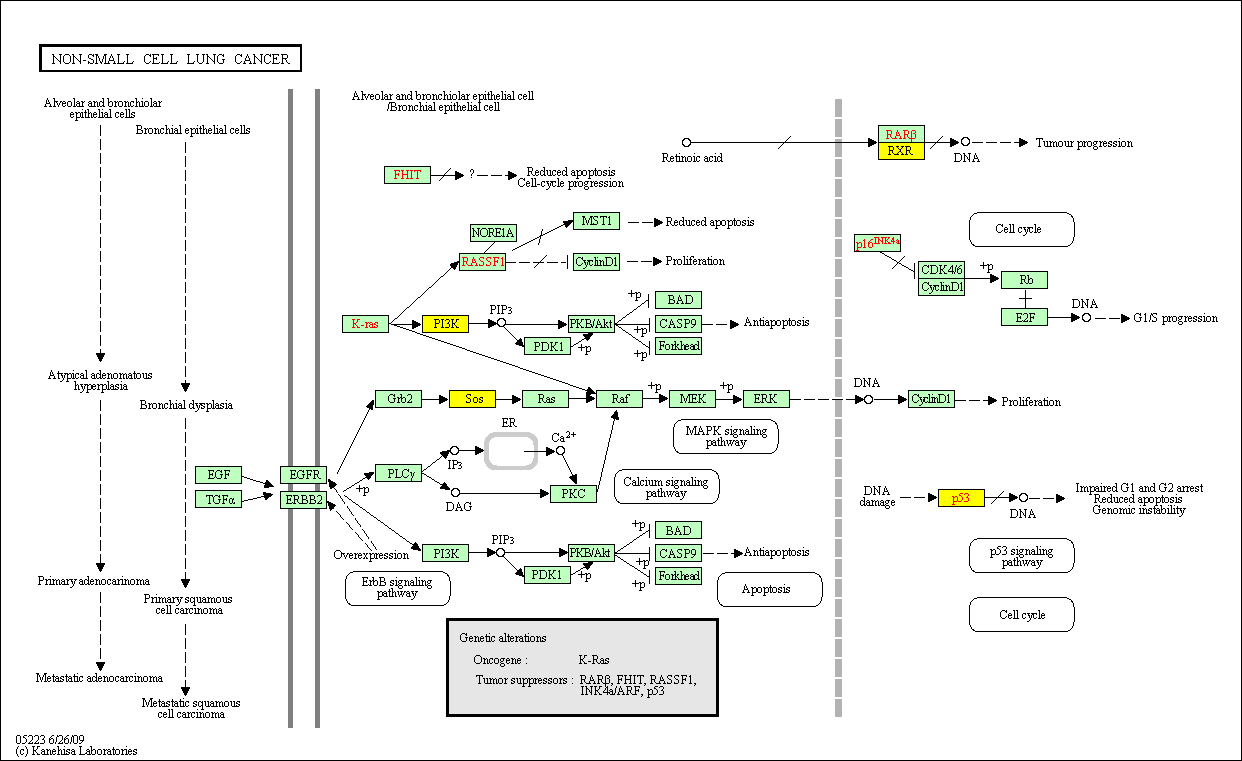
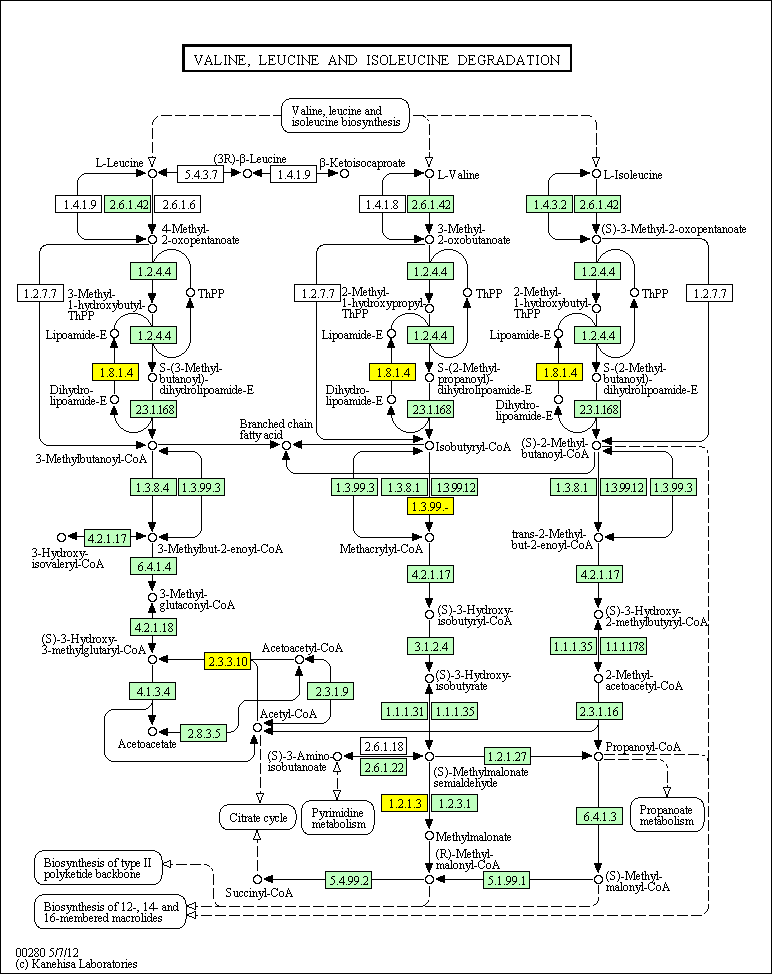
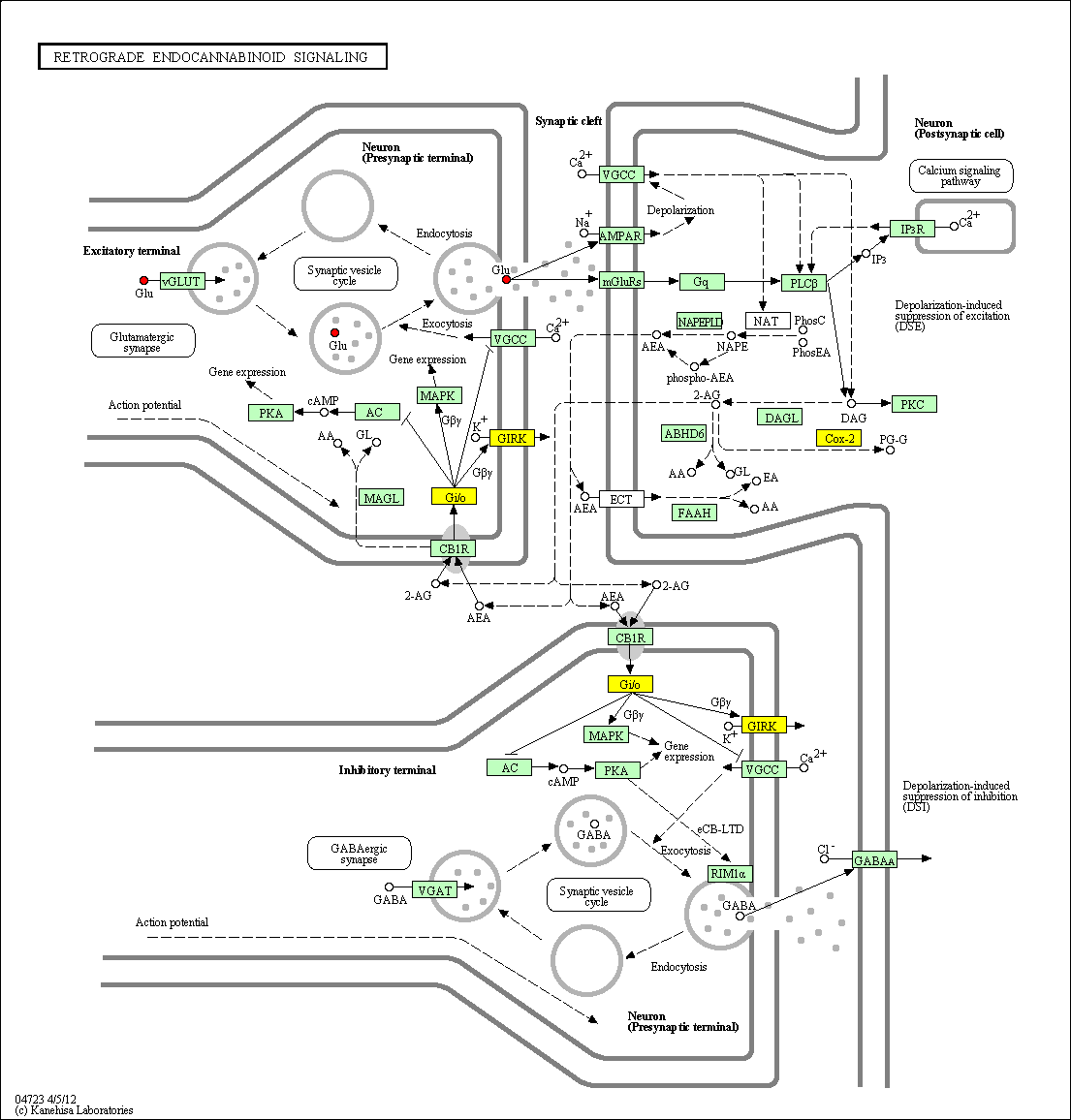
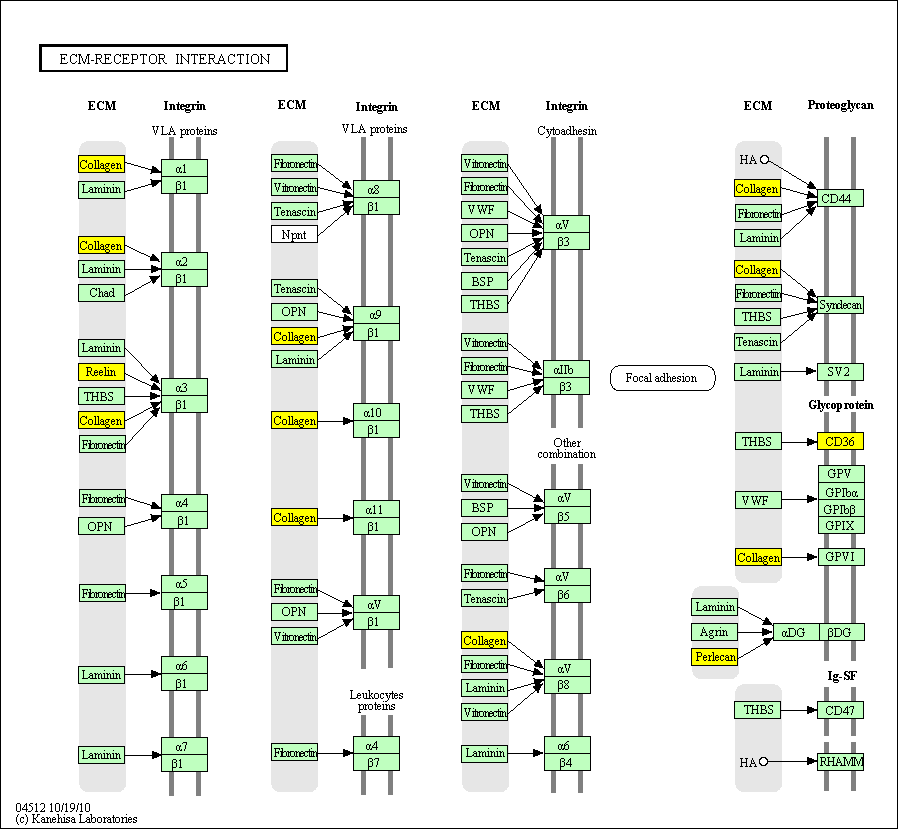
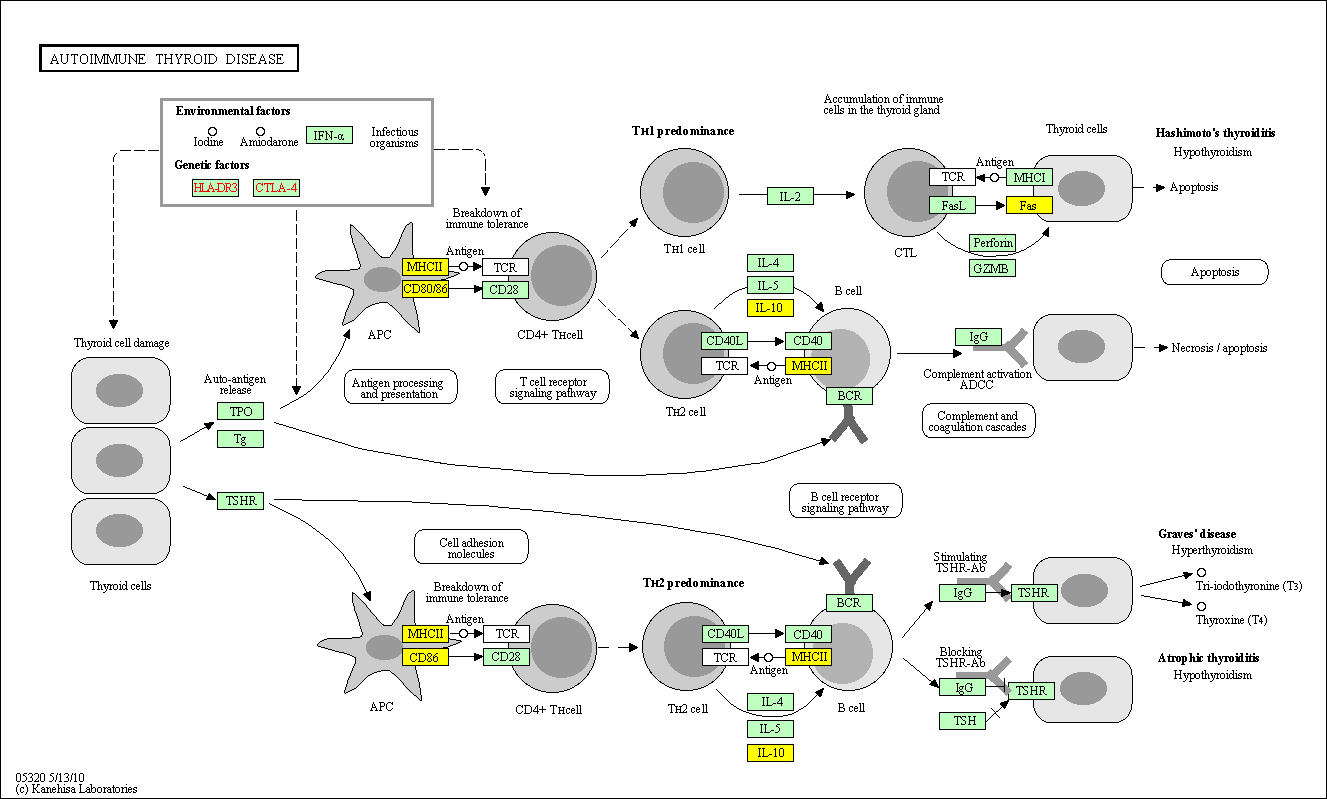
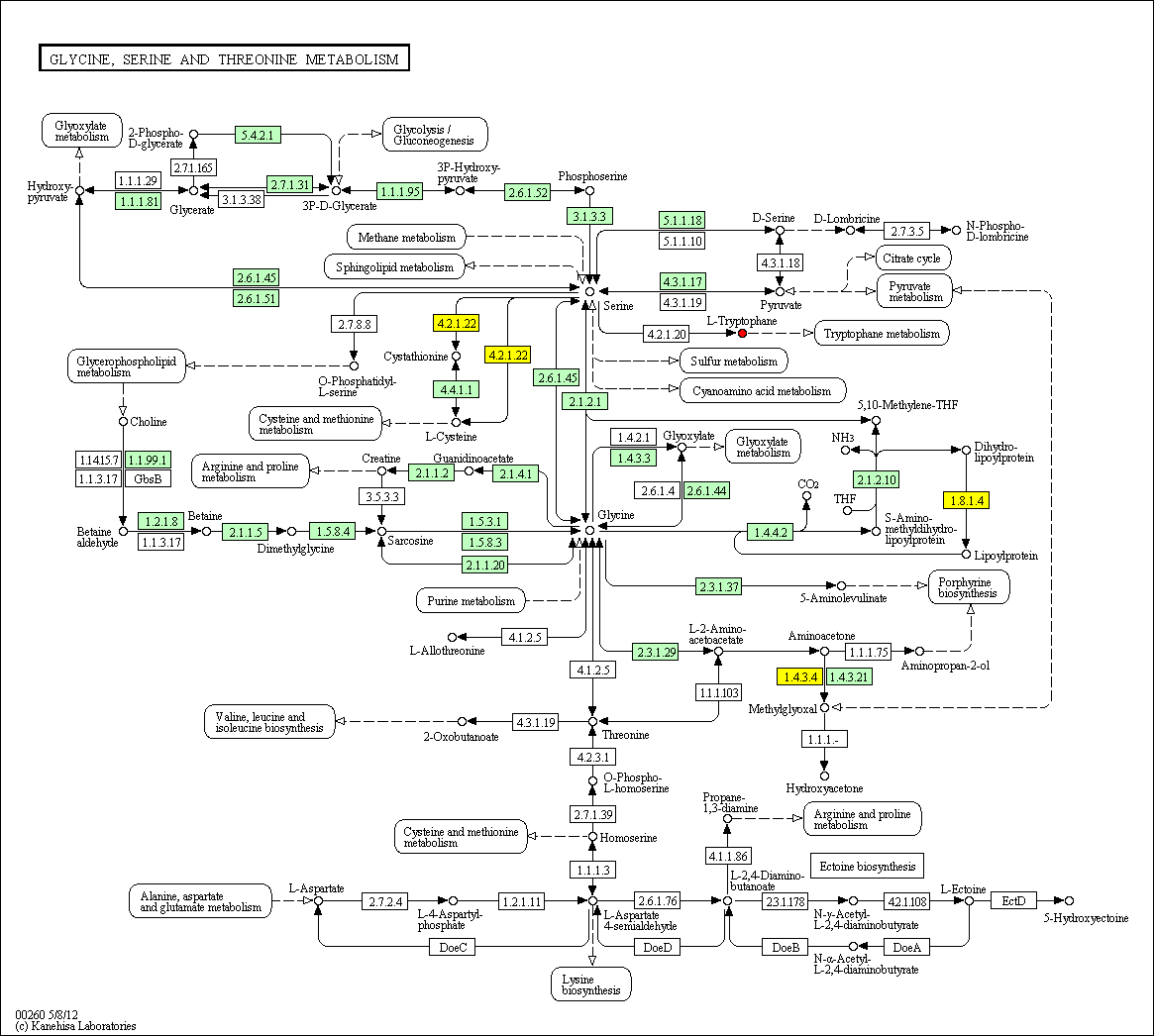
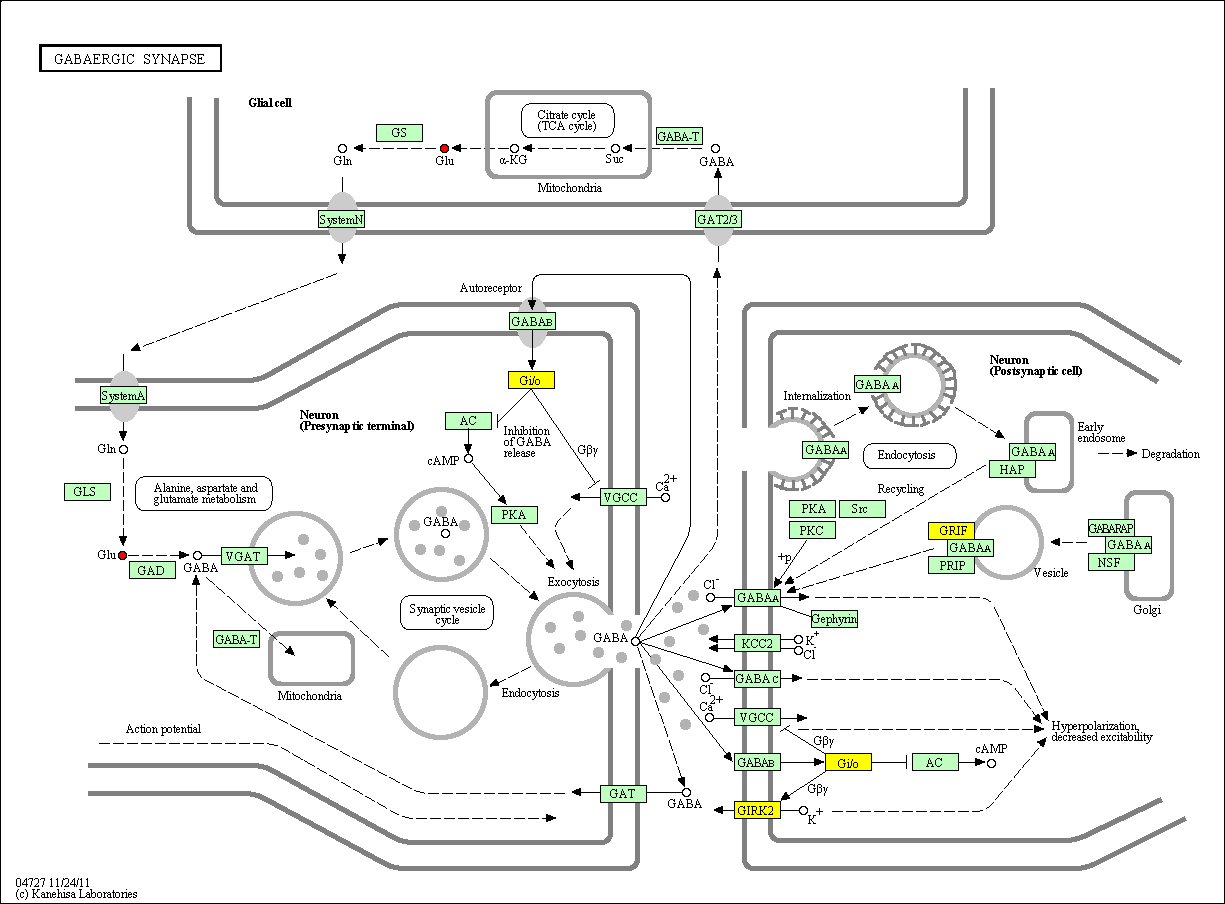
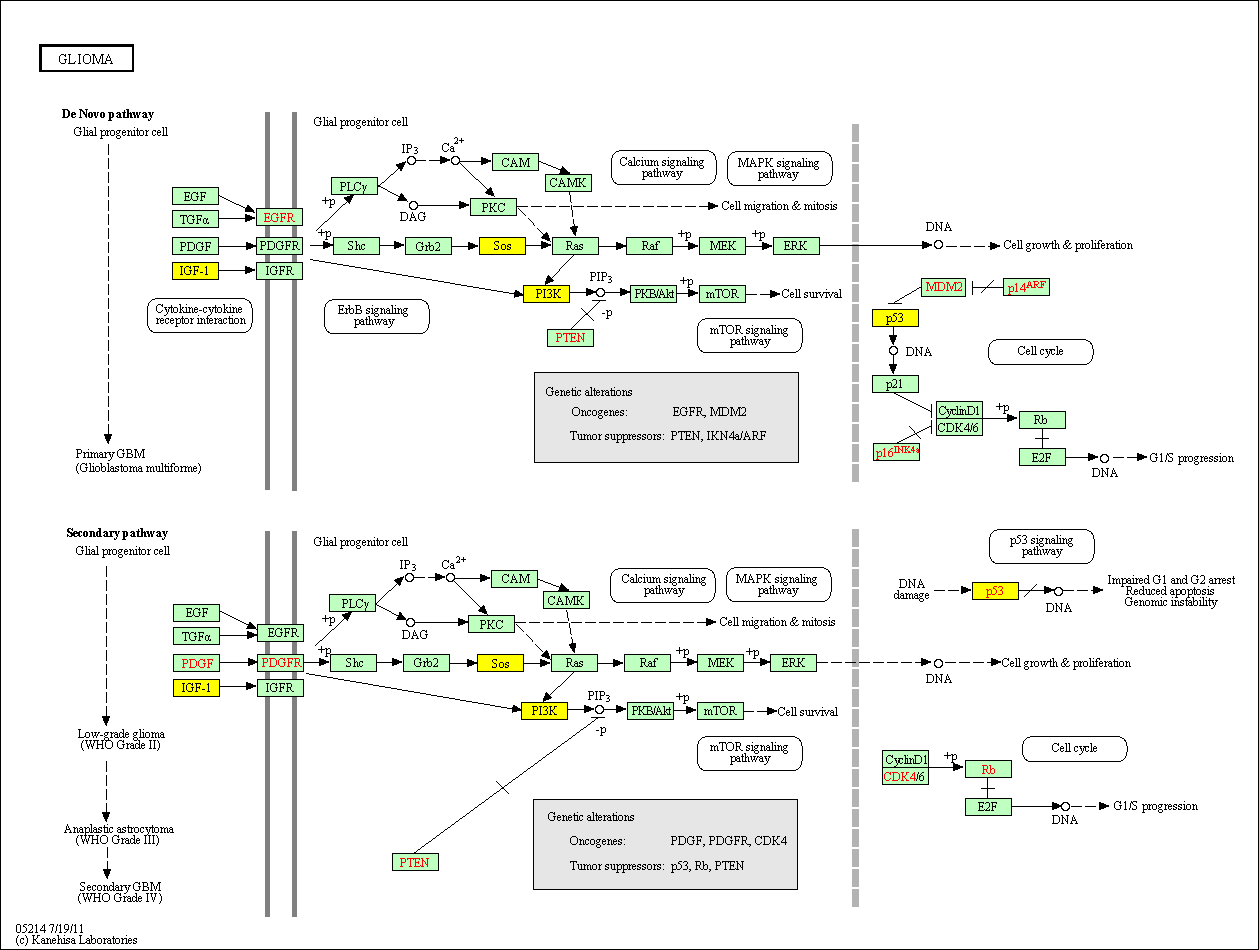
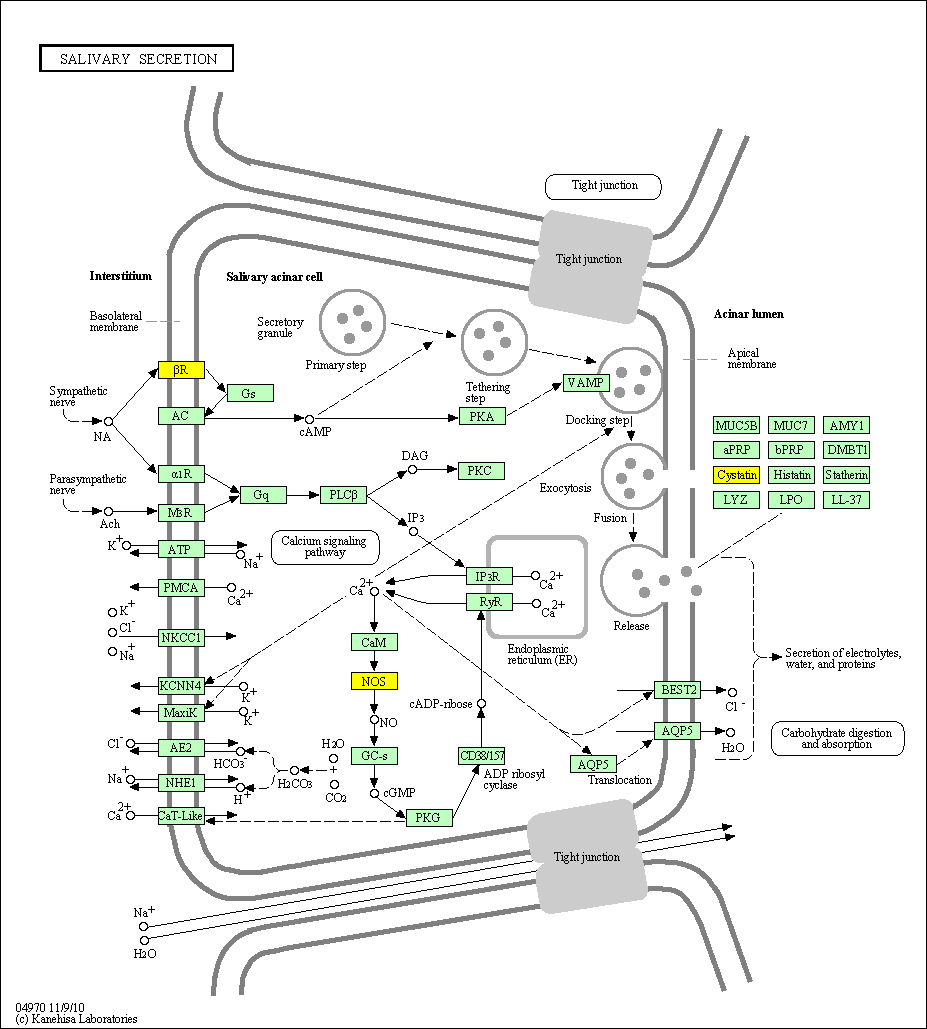
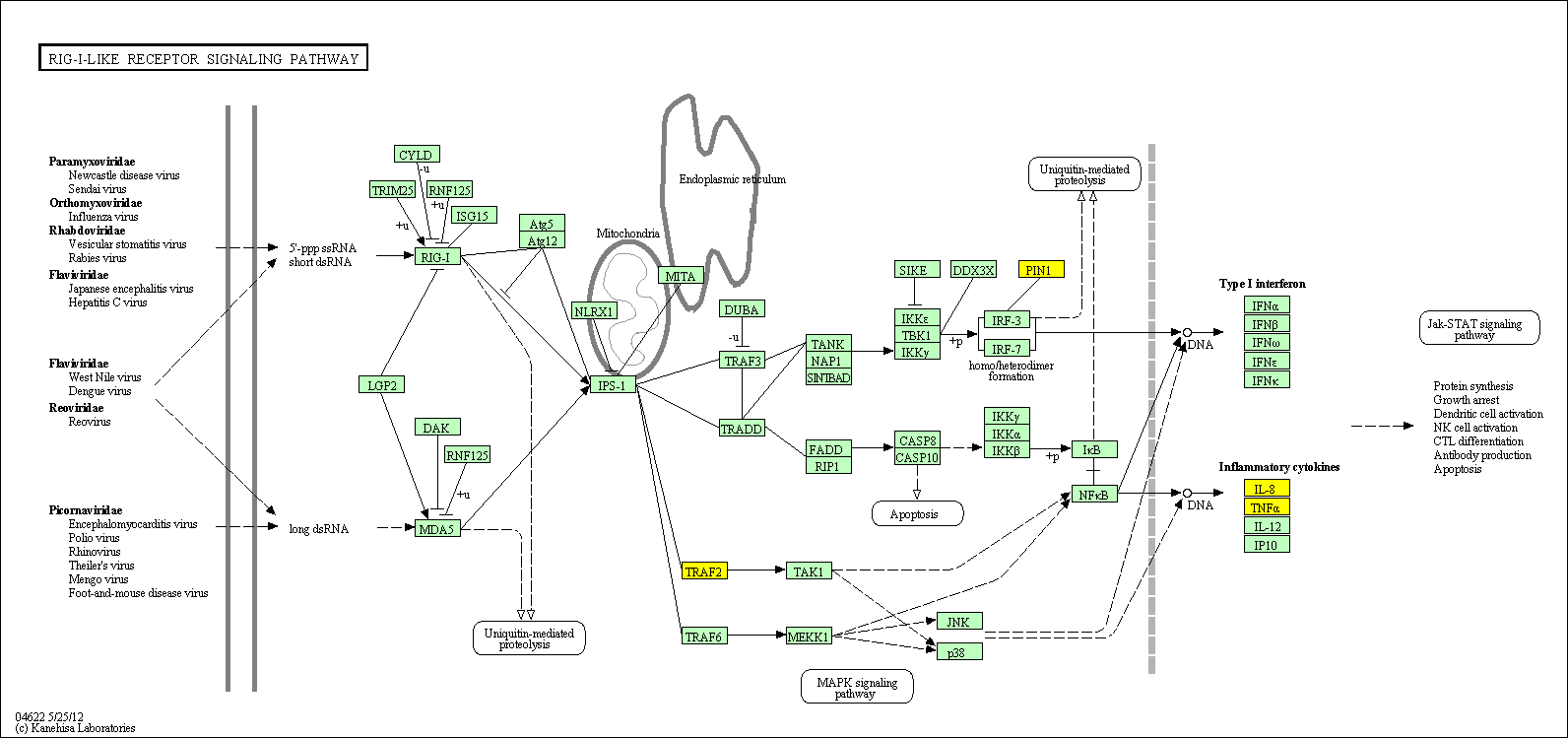
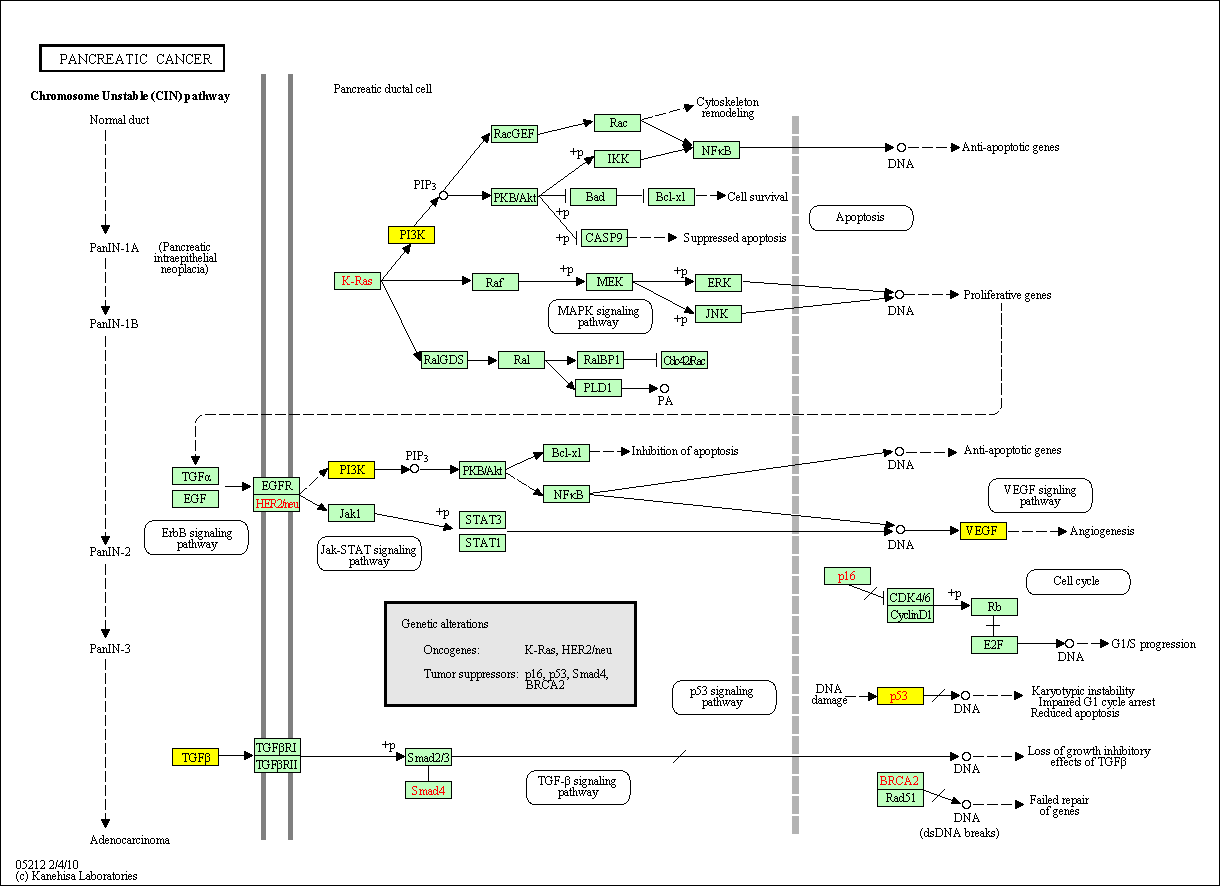
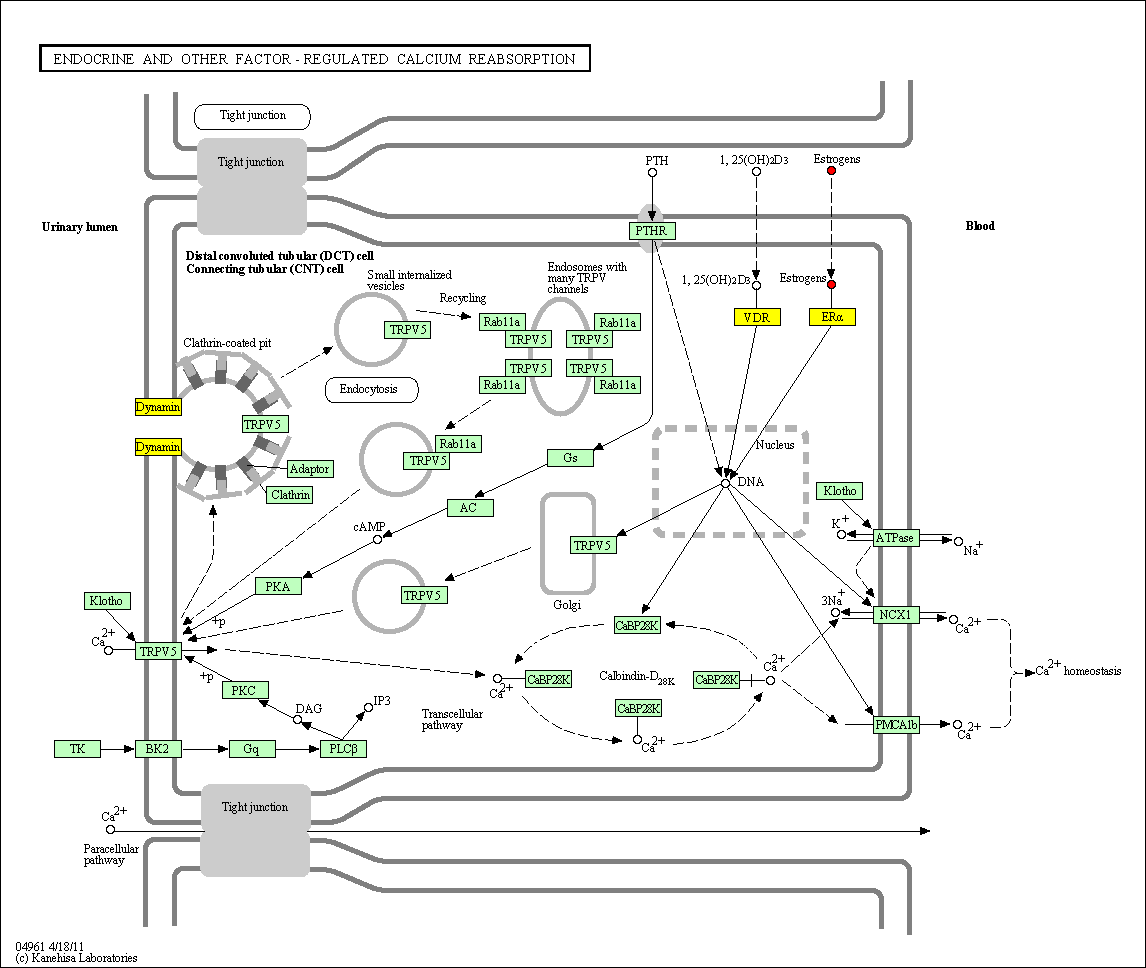
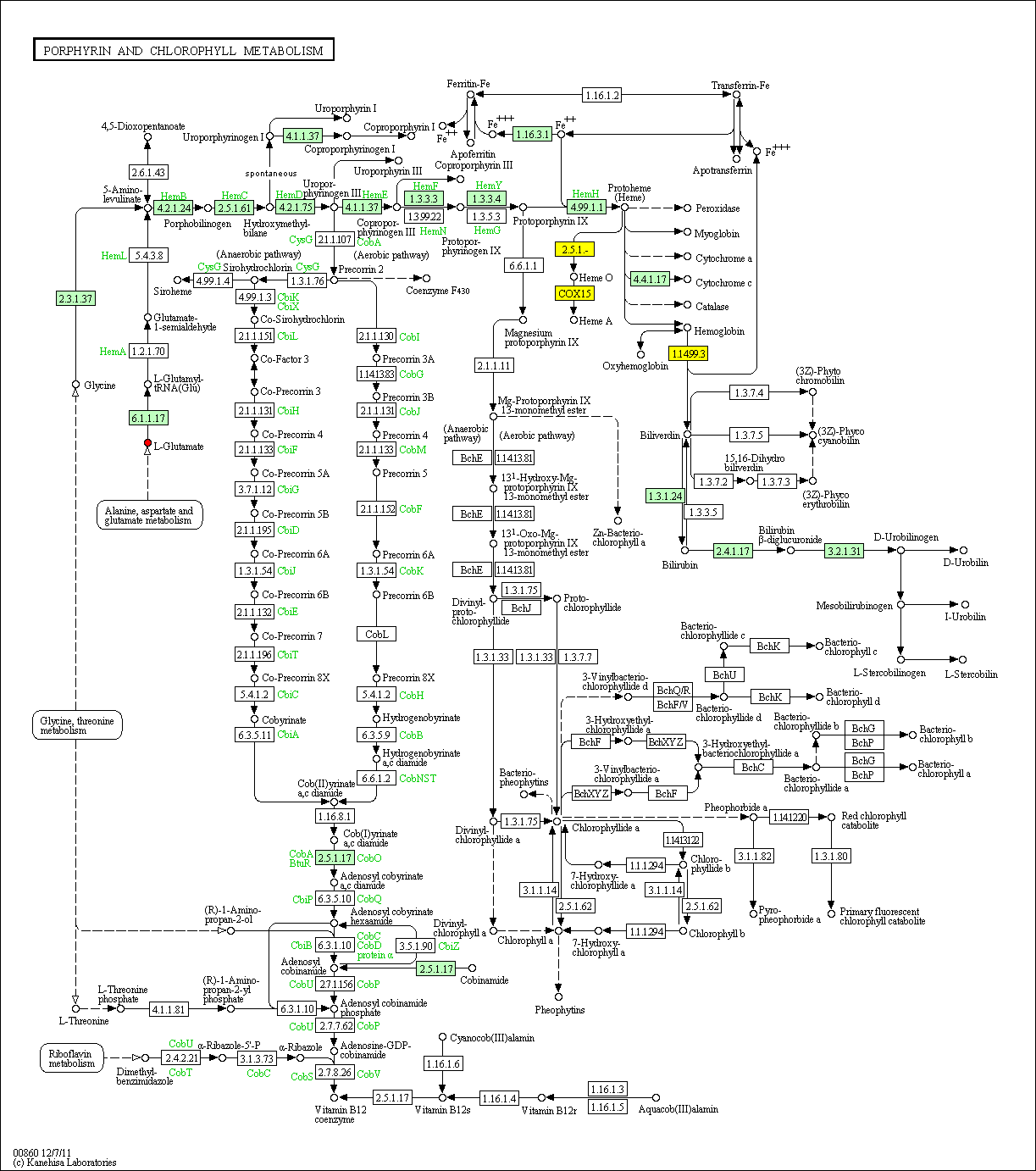
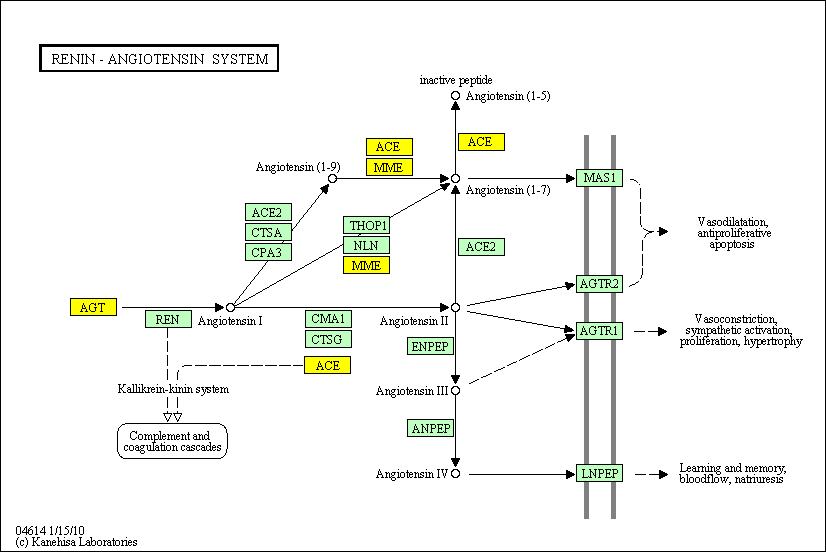
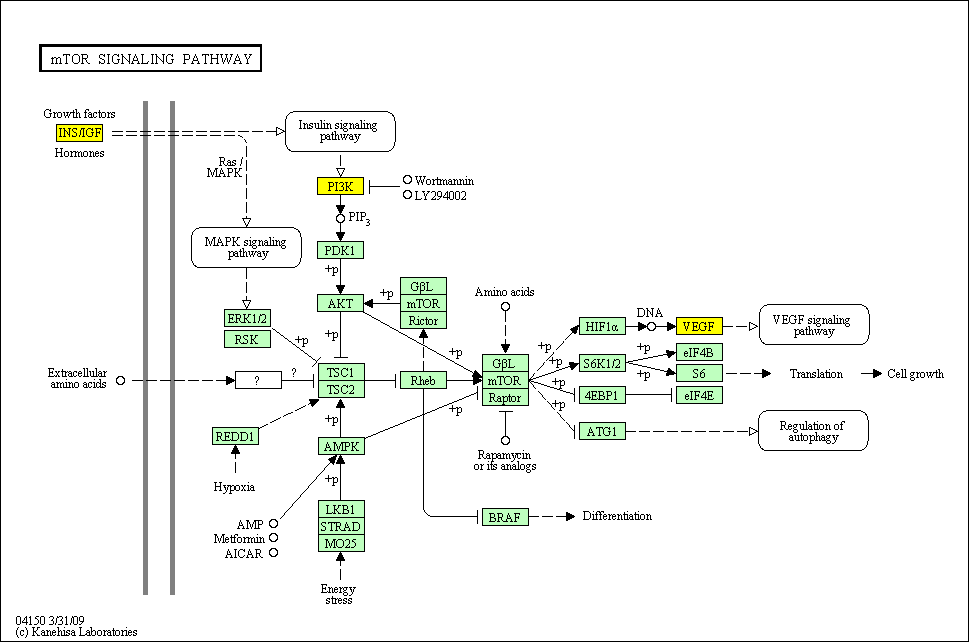
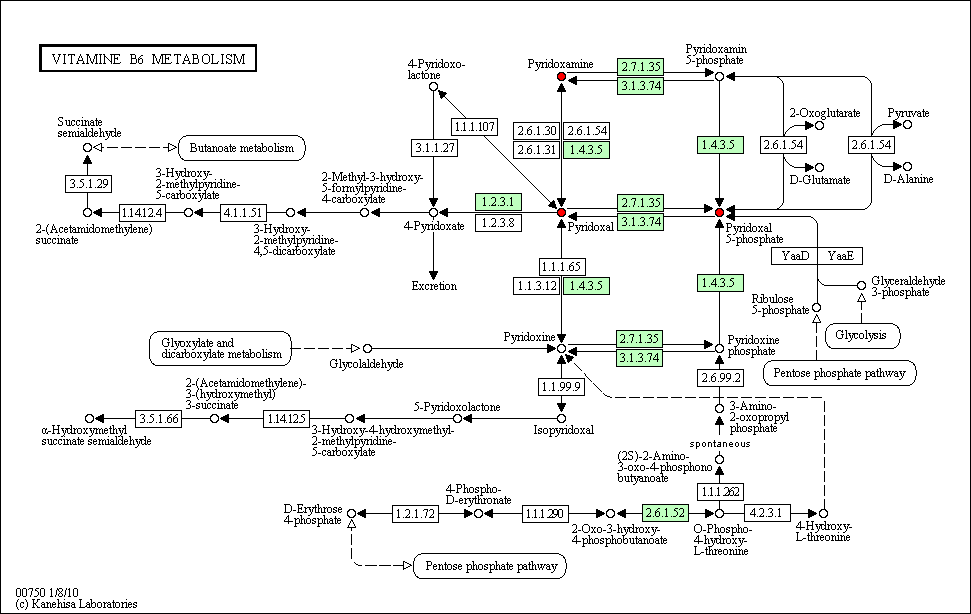
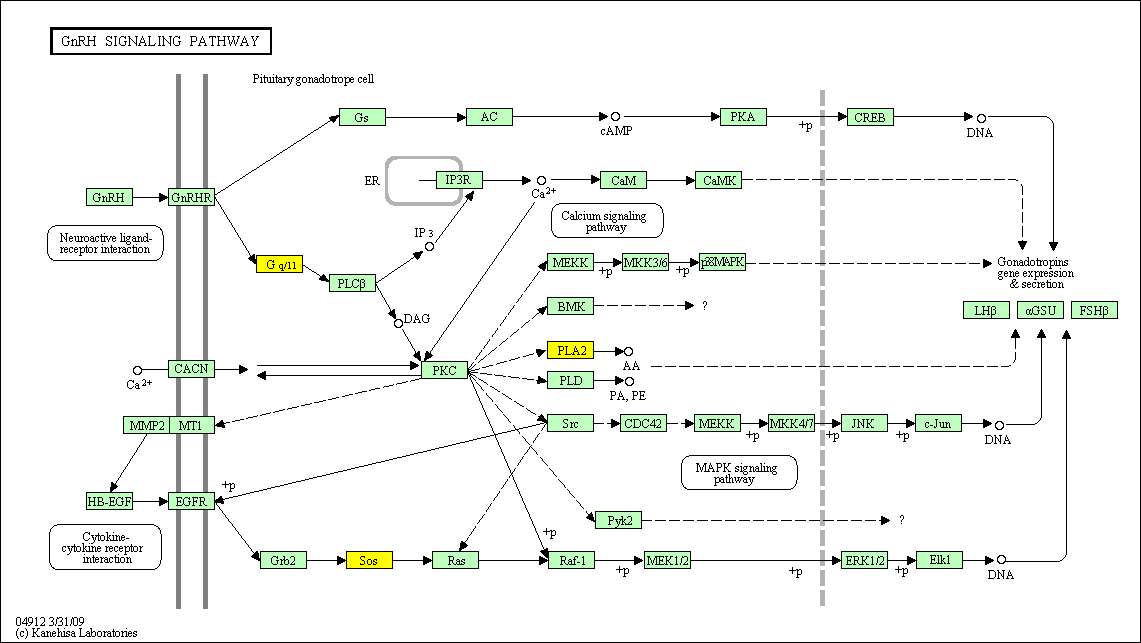
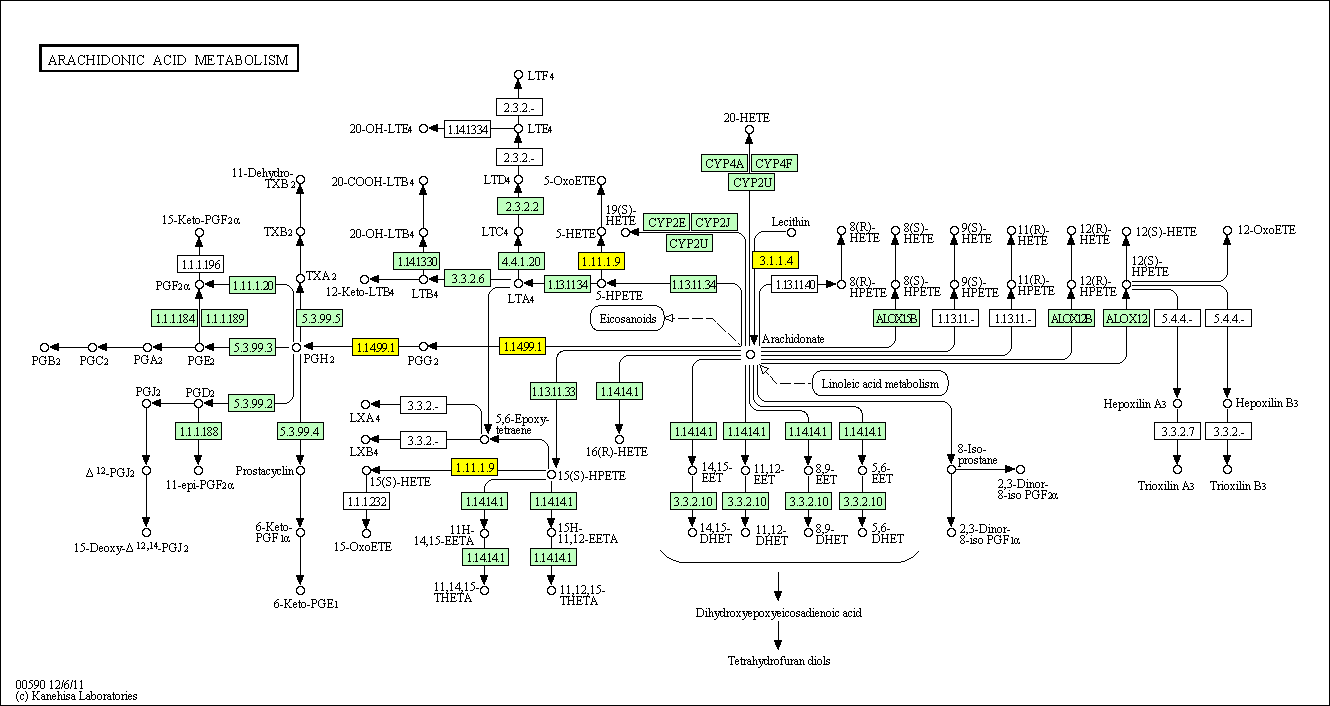
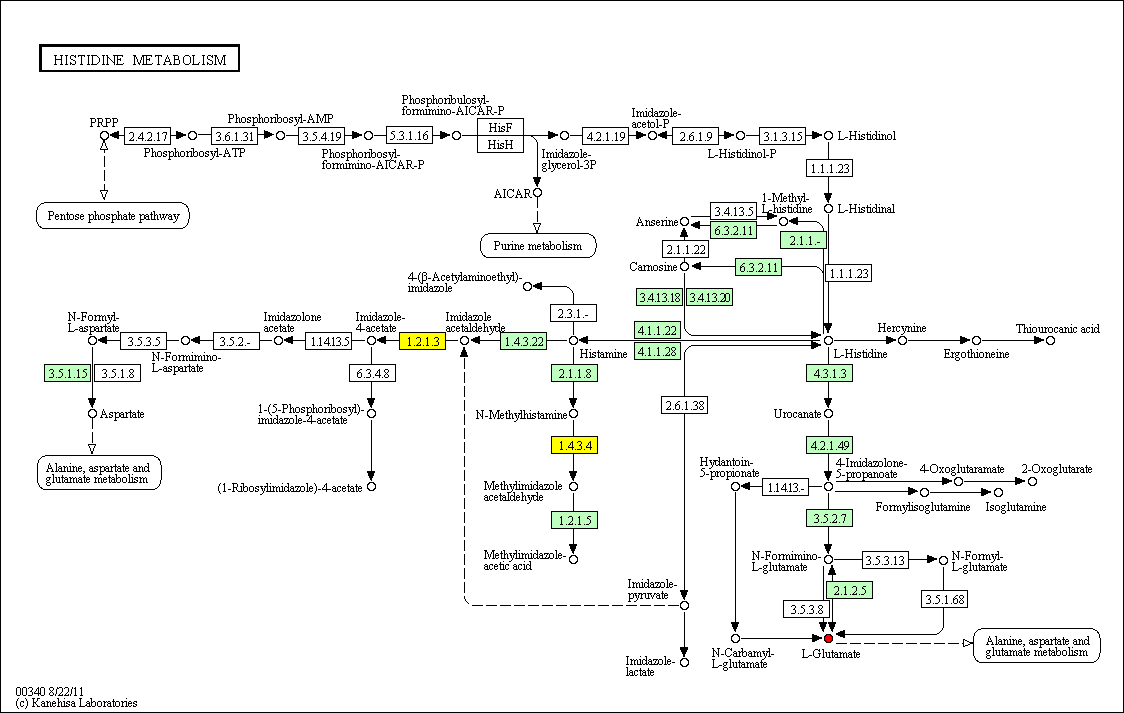
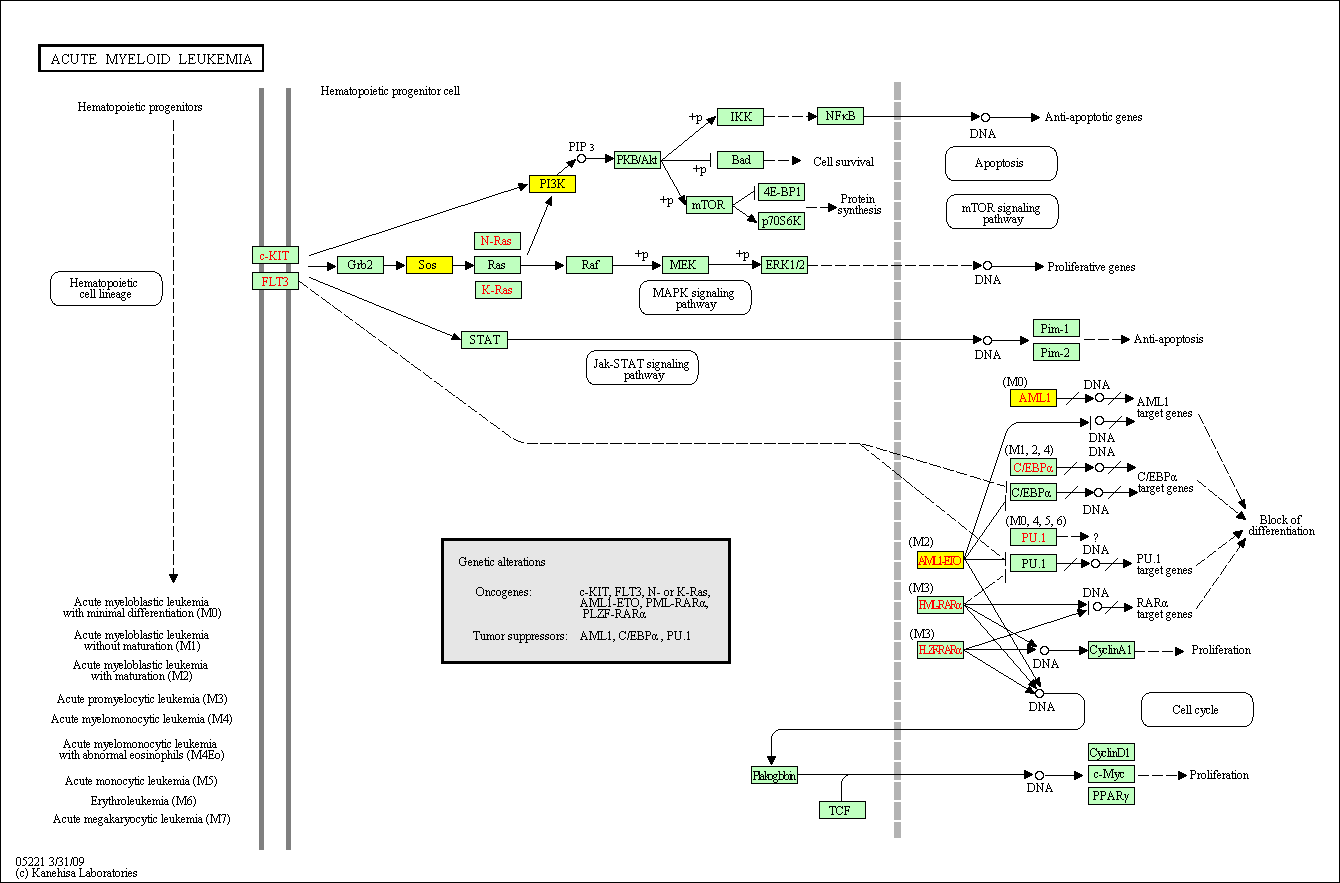
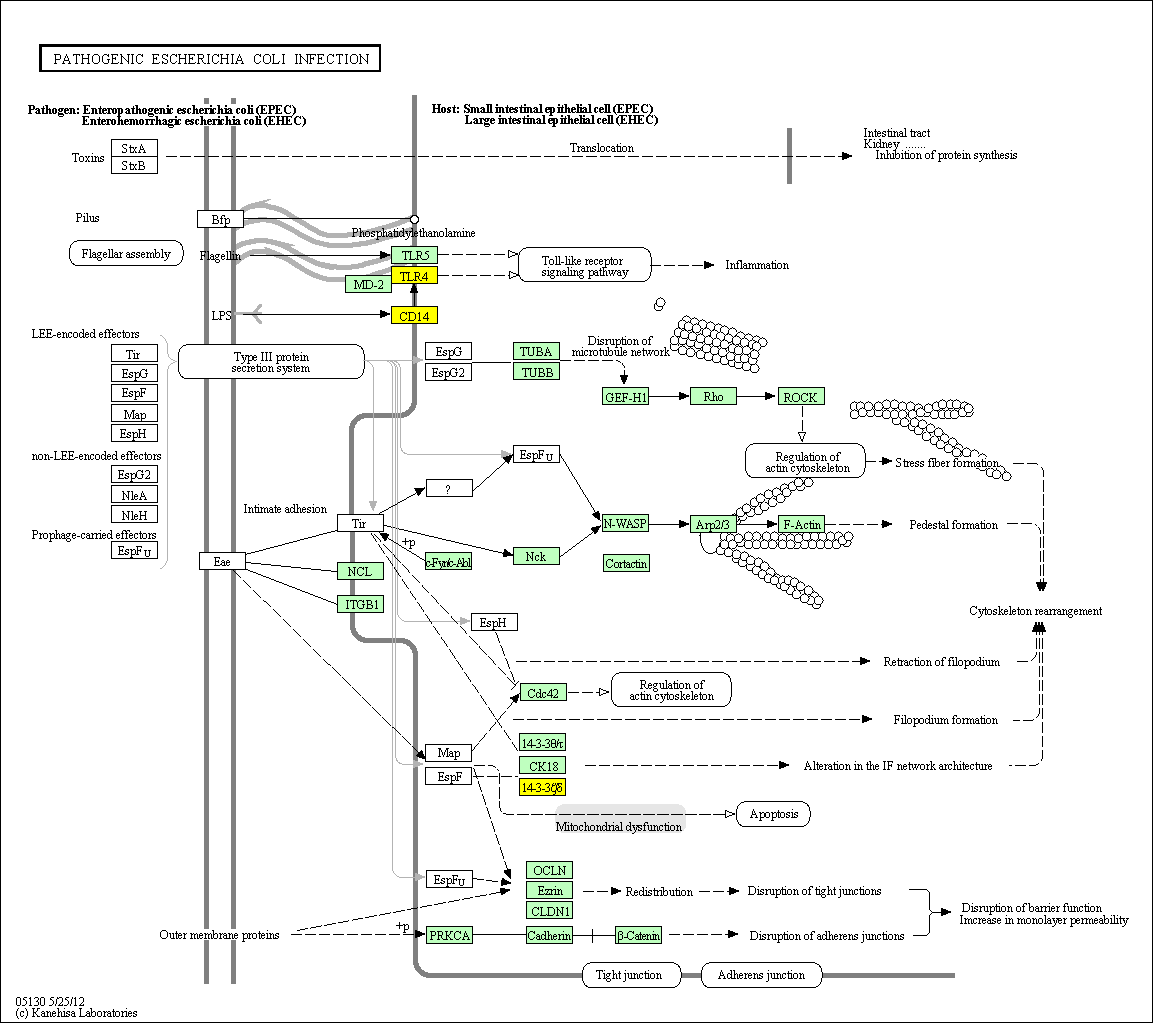
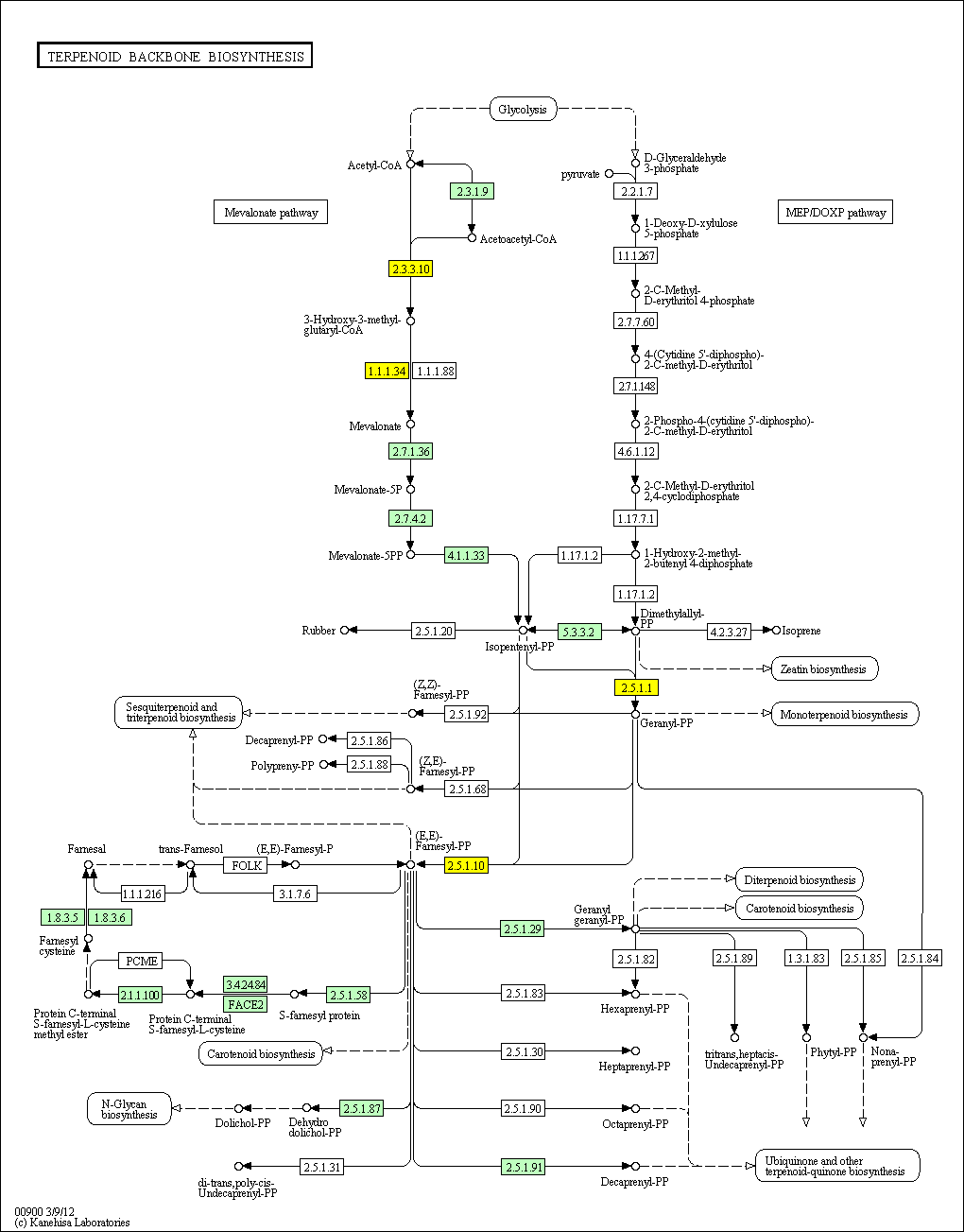
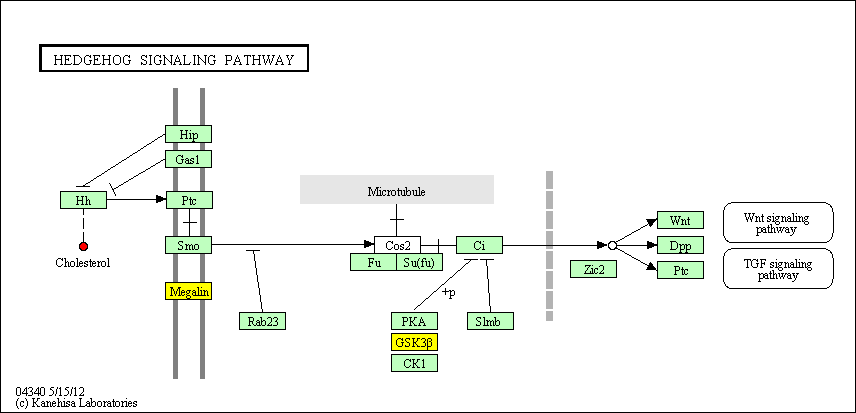
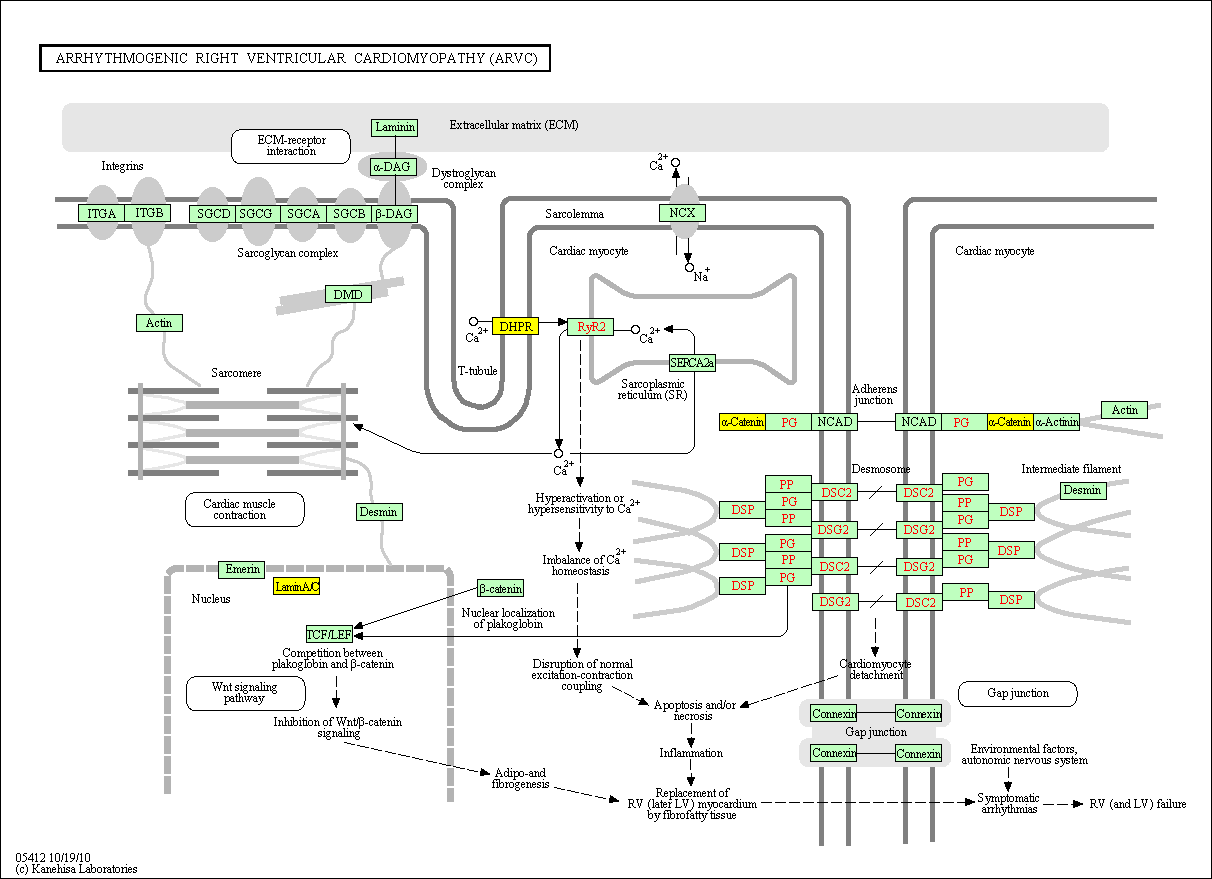
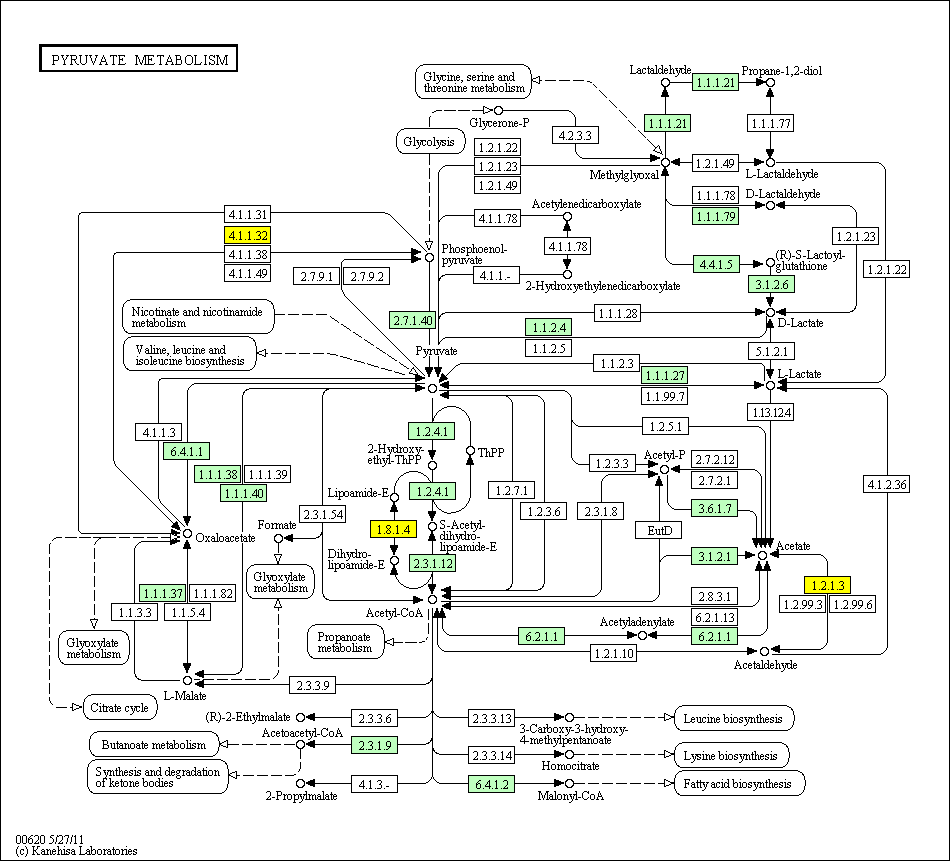
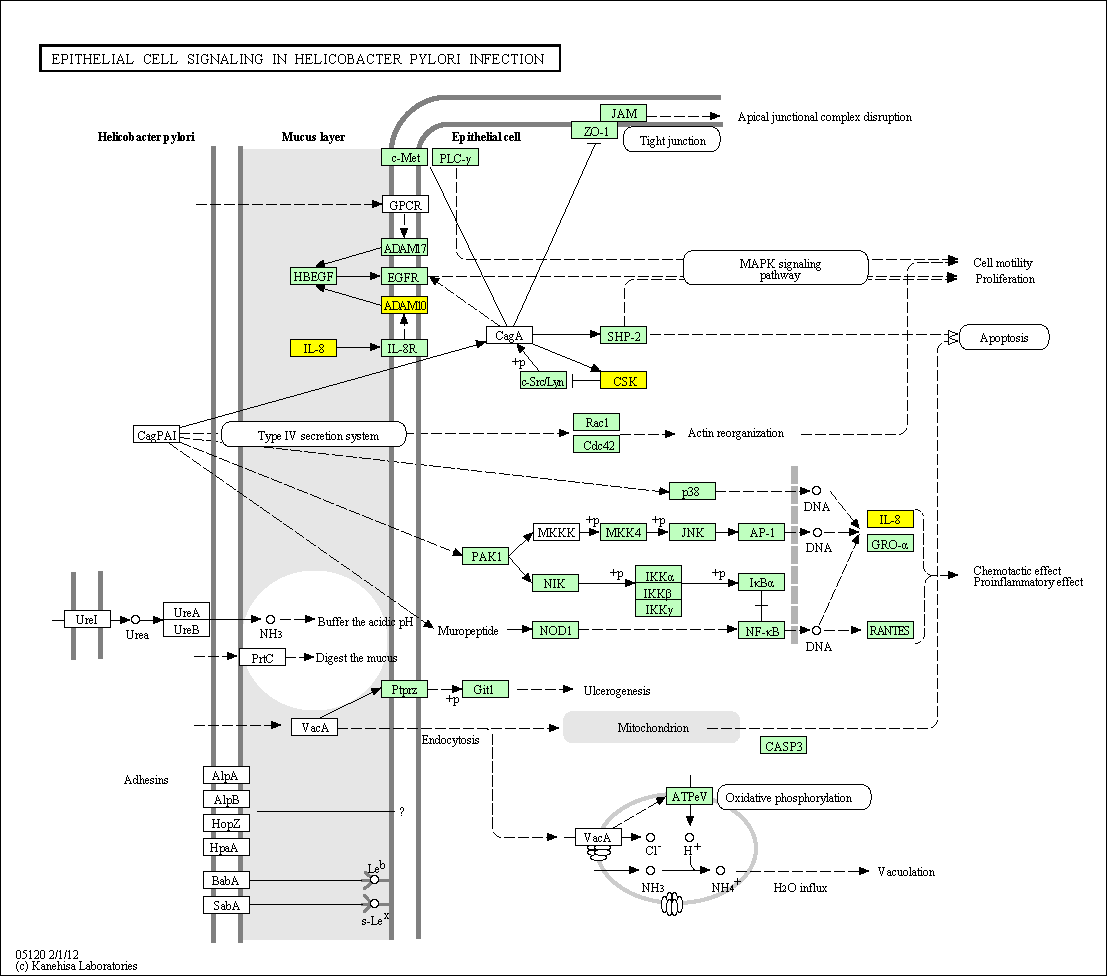
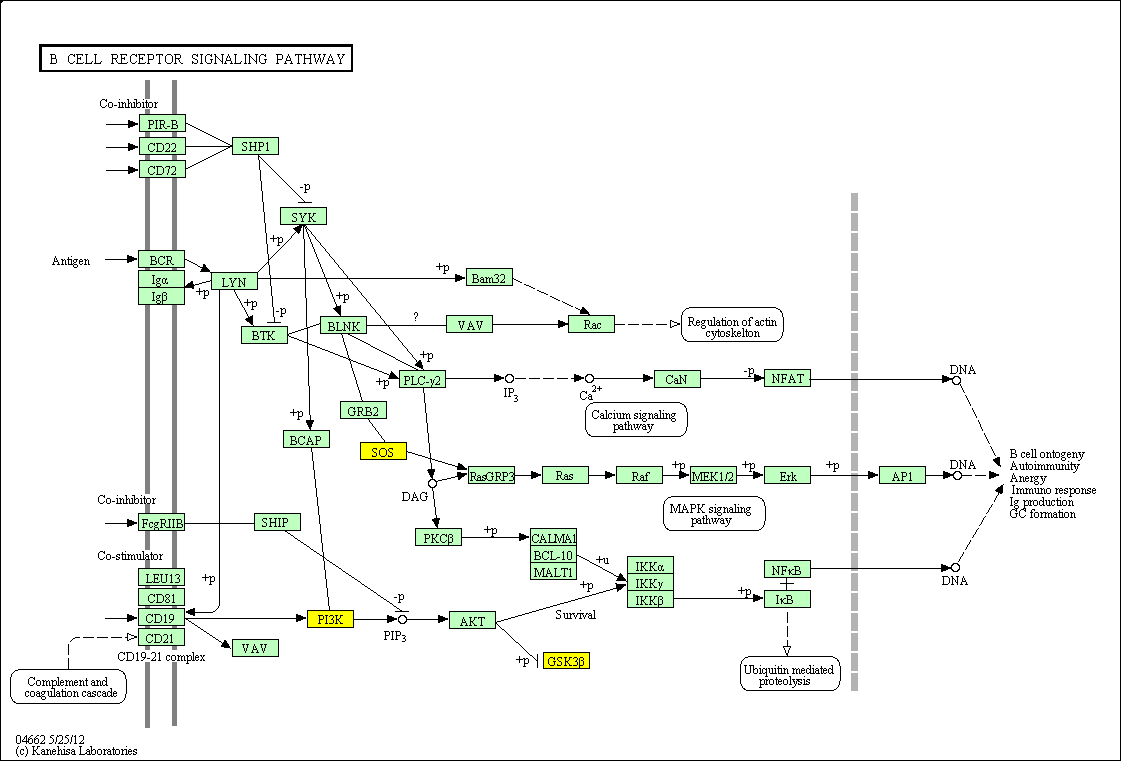
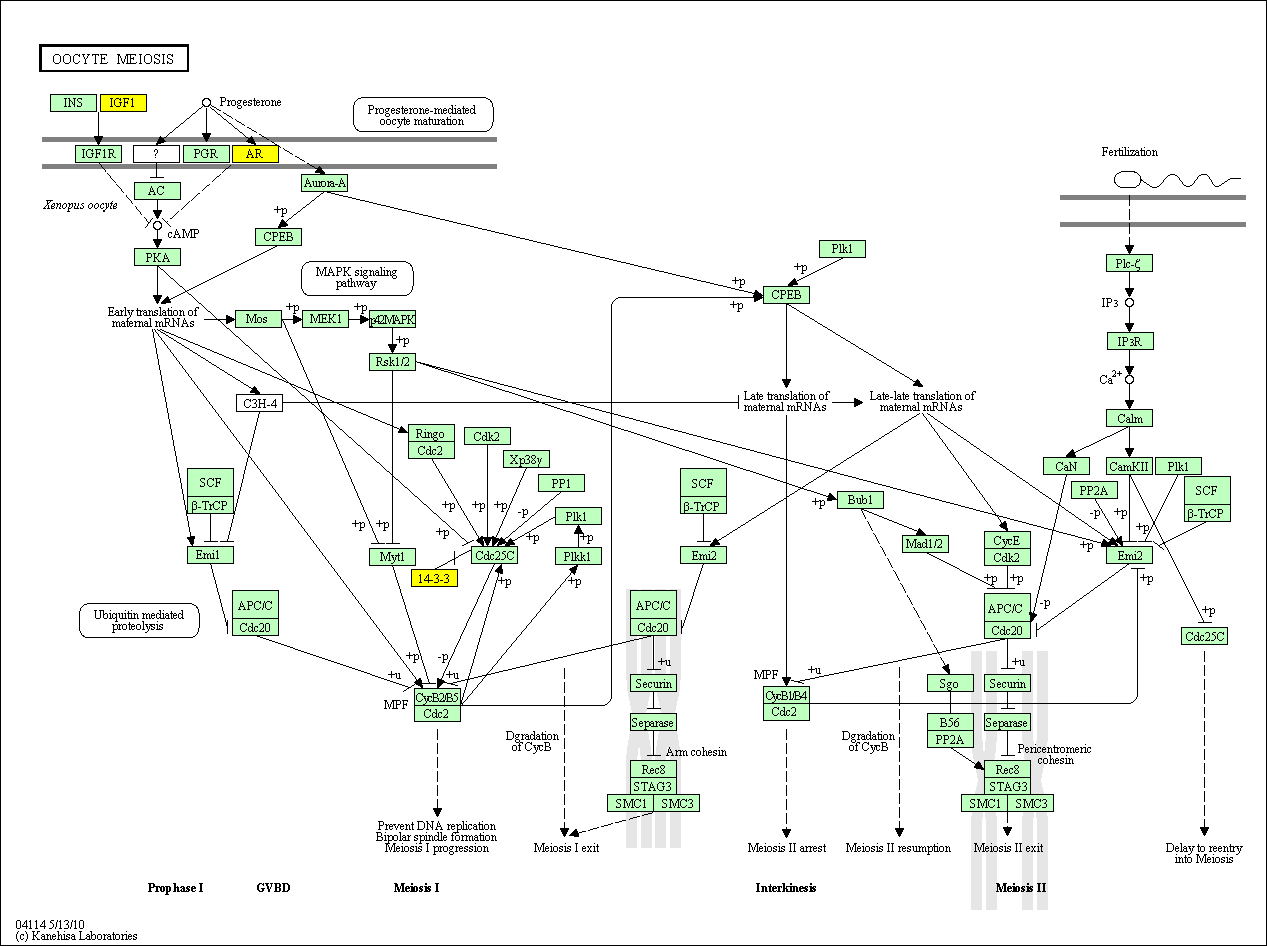
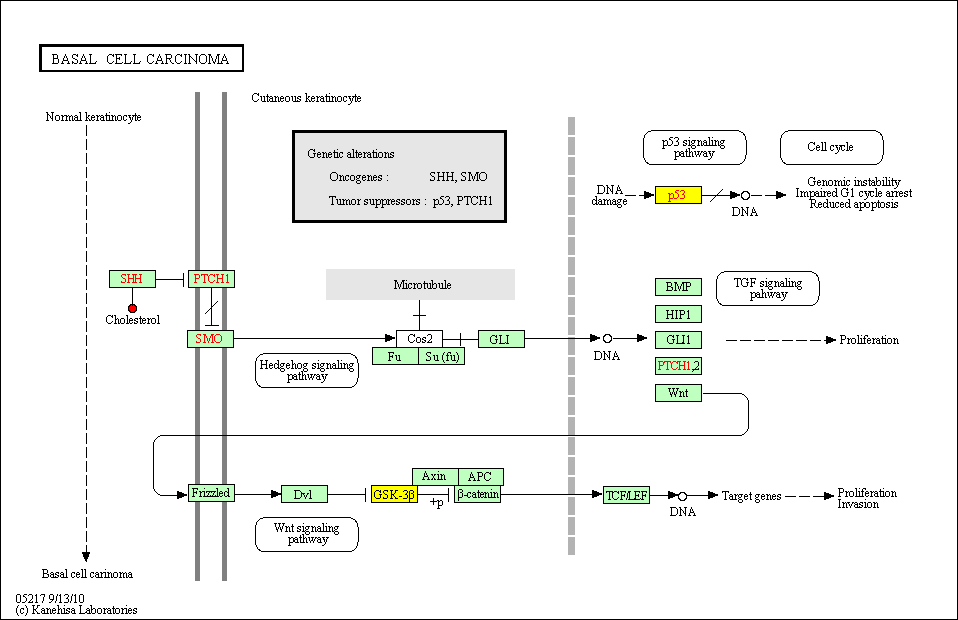
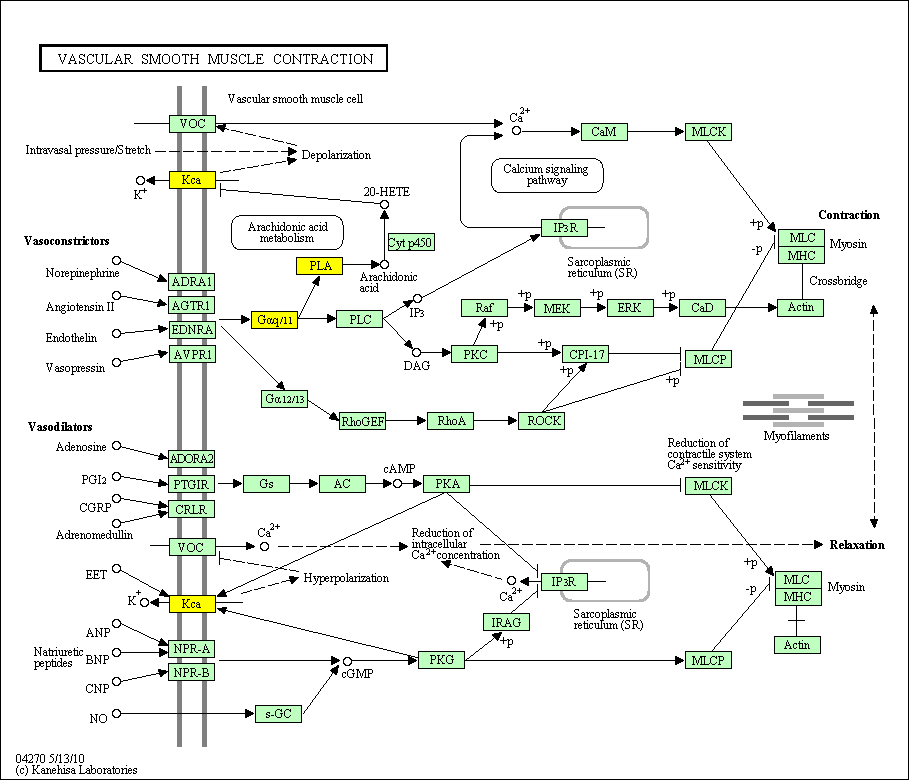
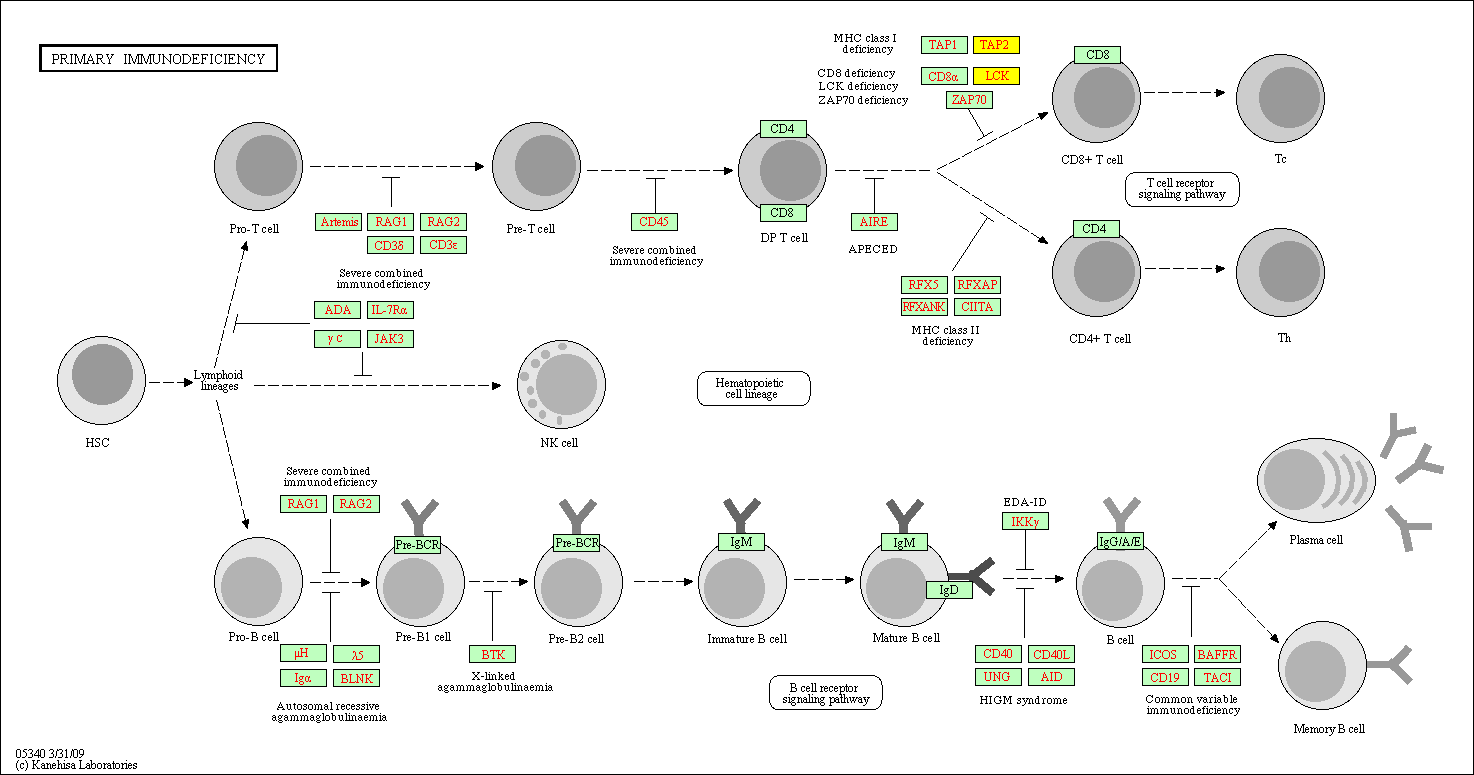
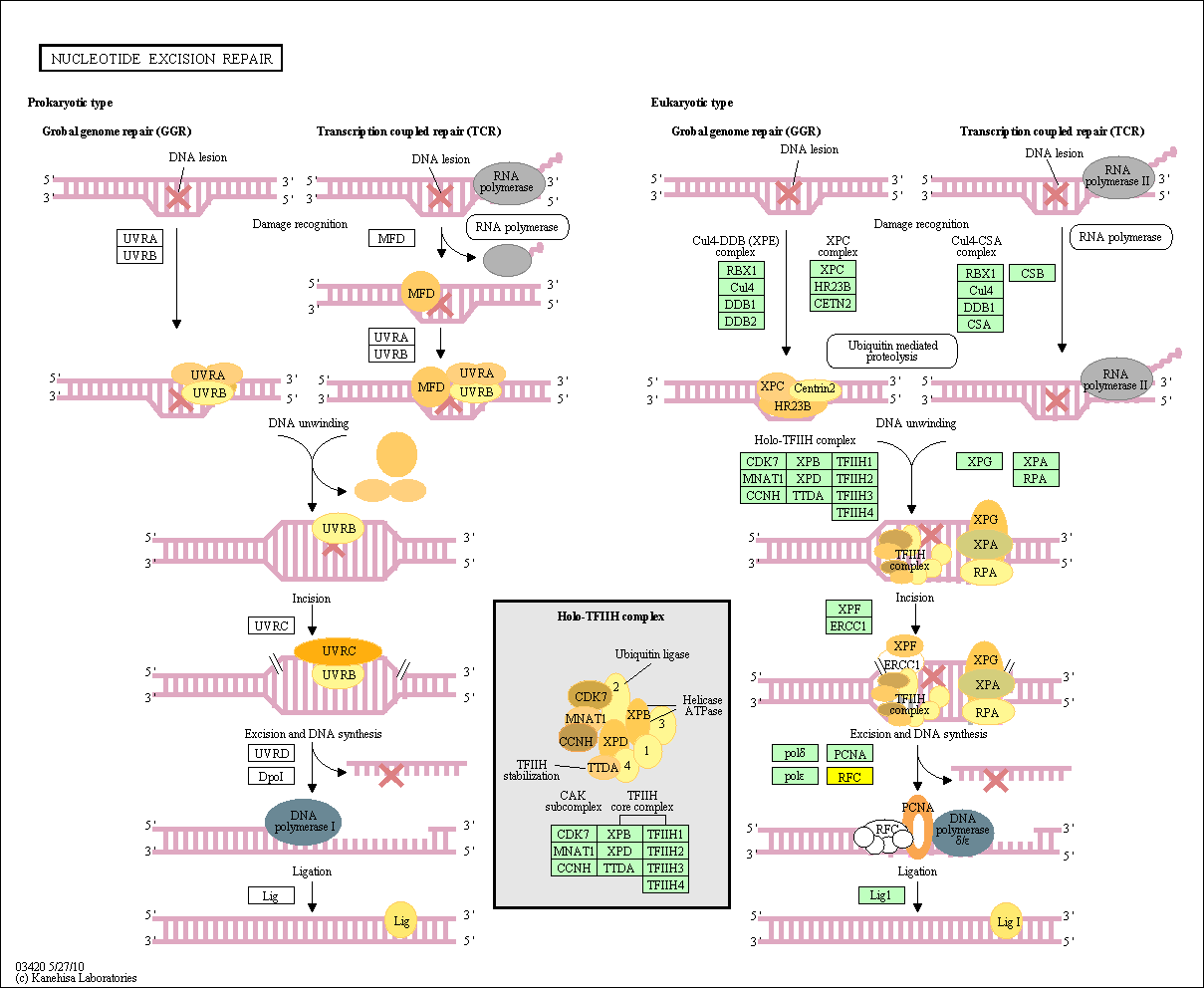
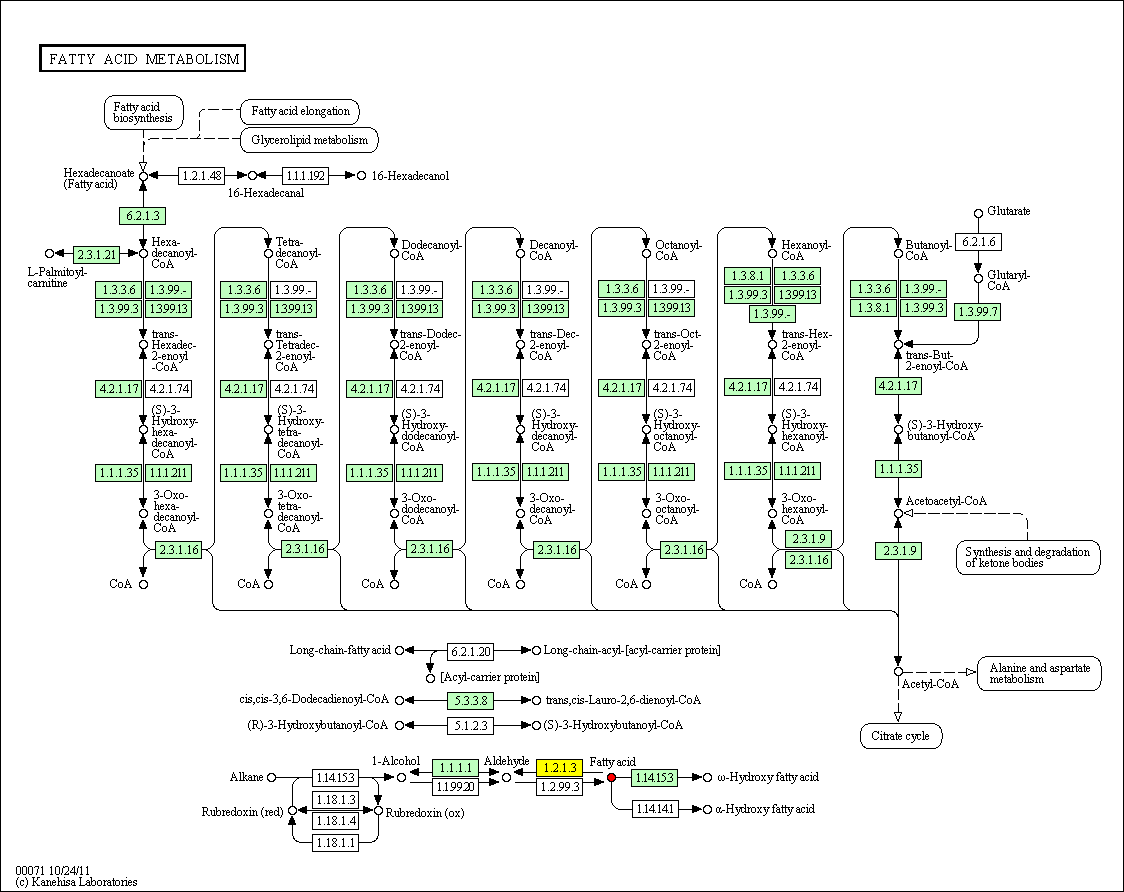
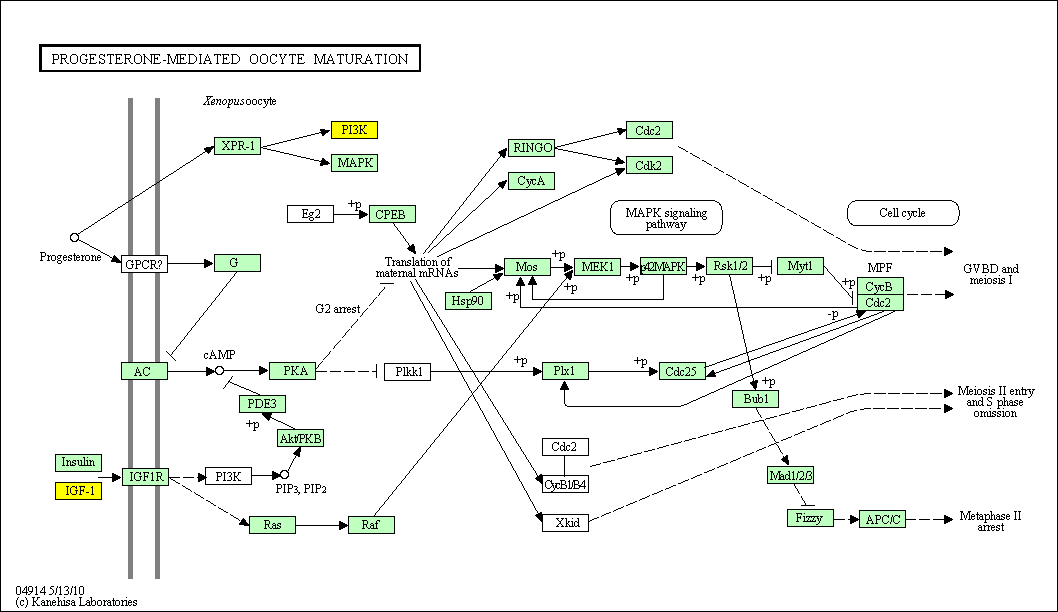
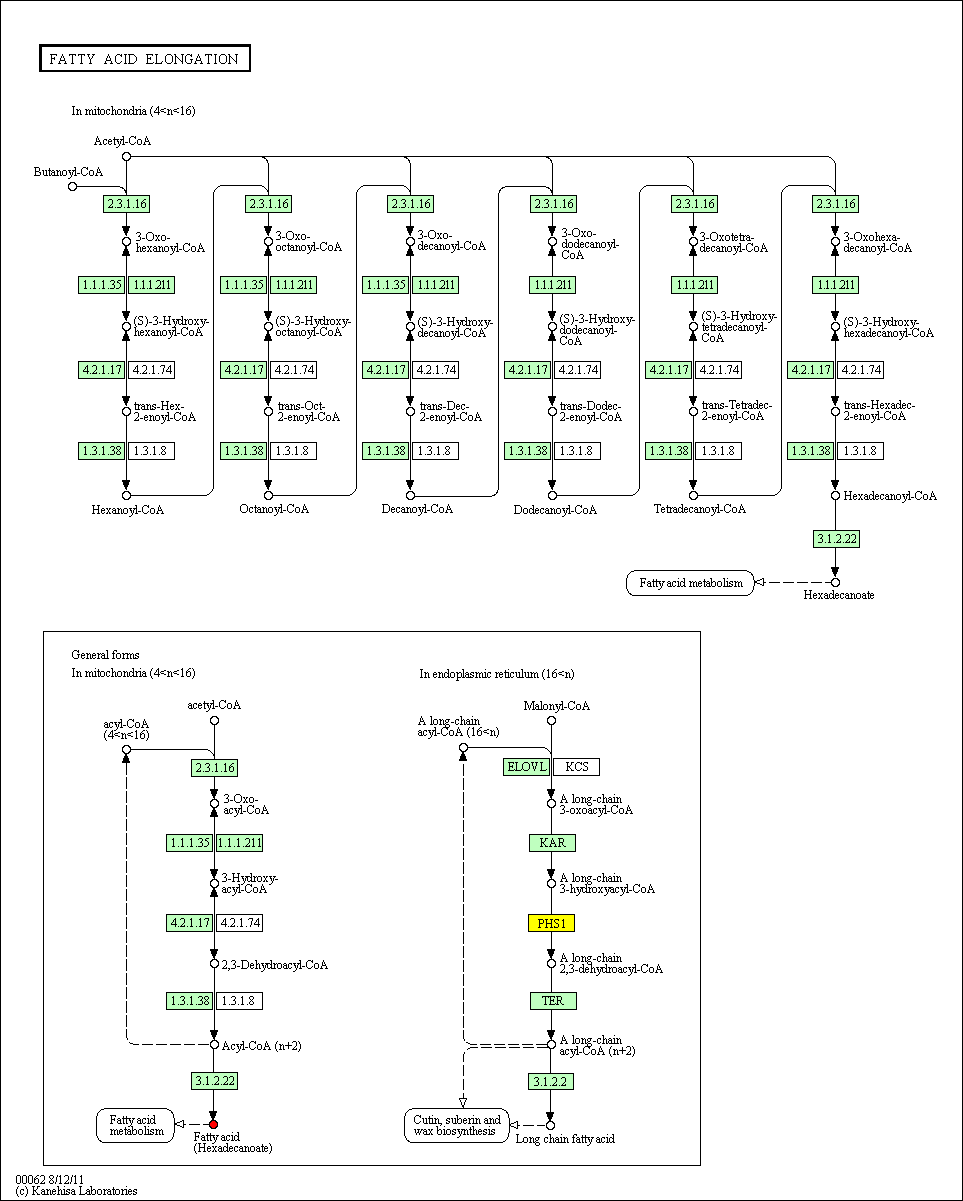
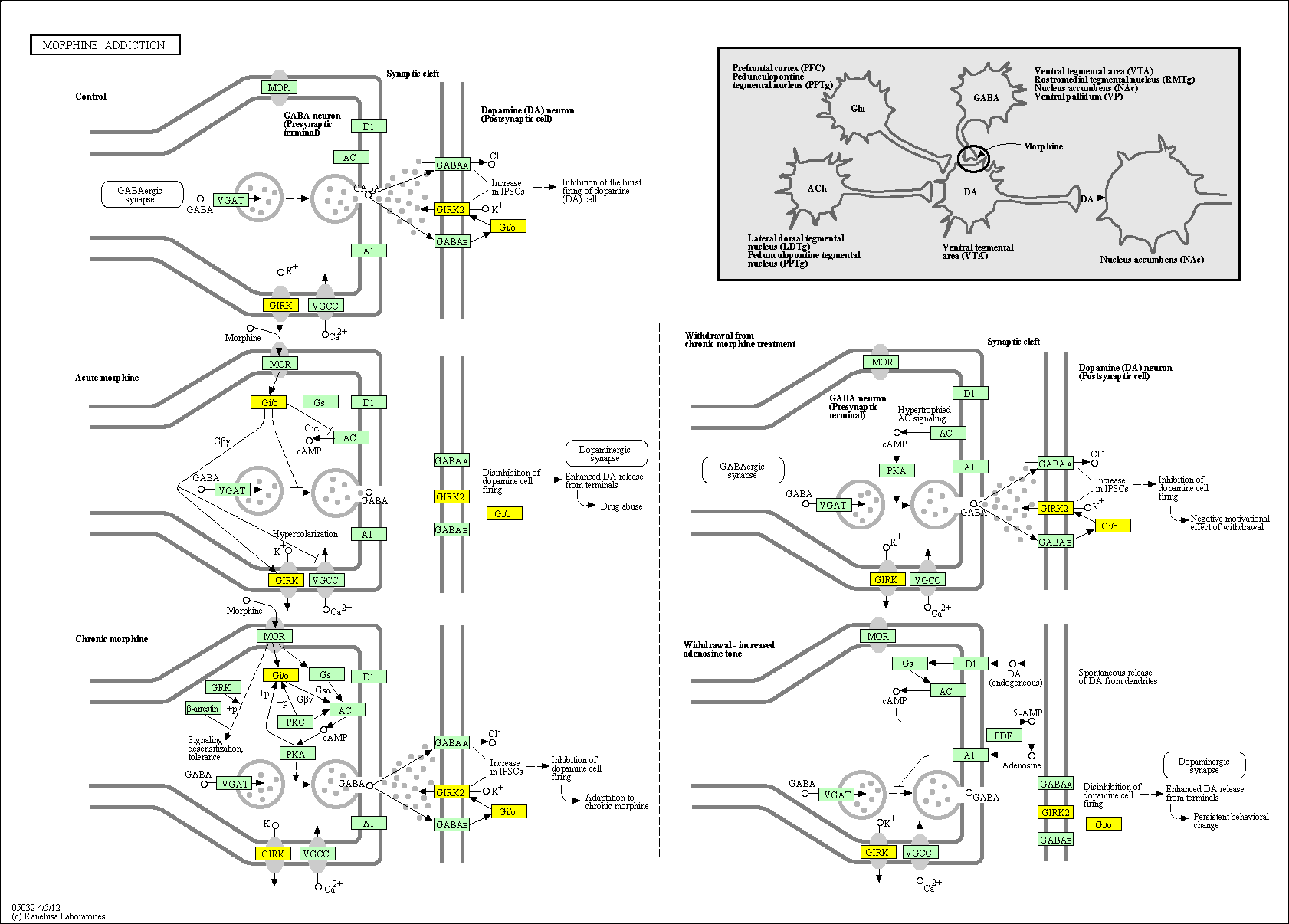
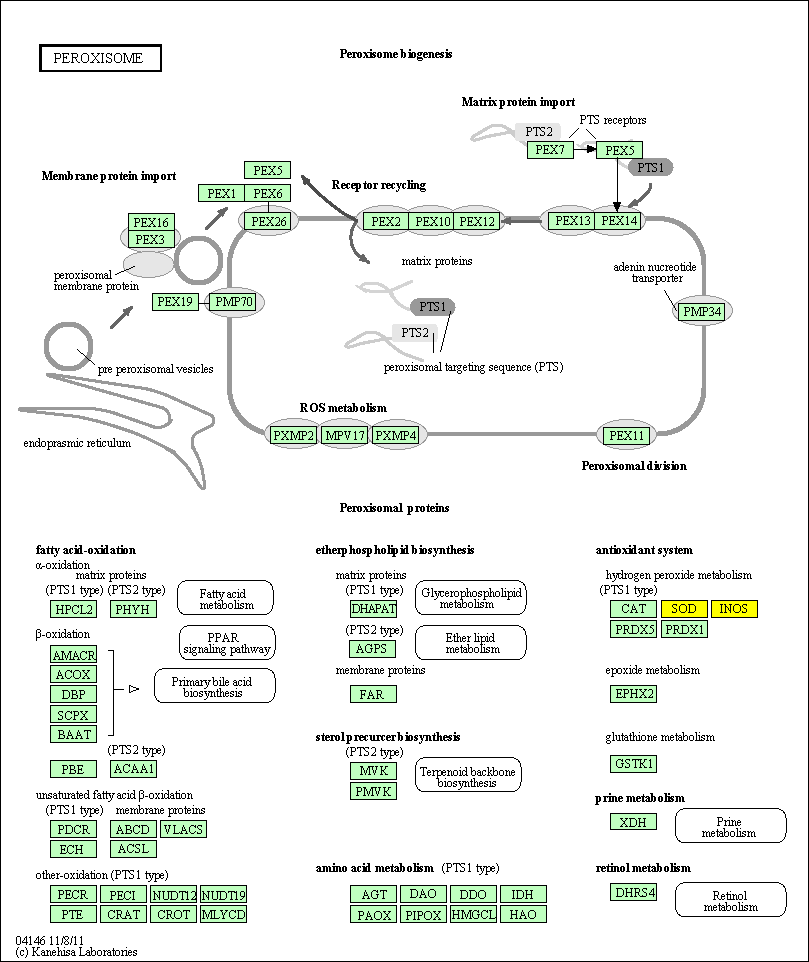 hsa:4843
hsa:4843